- Selecting attributes from a data frame
- subsetting rows
- Filtering / extracting / subsetting data frames based on attribute value
- Filtering for rows where at least one column is missing
- Filtering for rows where all columns have values
- Exploring data
- Prevent garbage characters when using read.csv on data exported from SQL
- List functions exported by a loaded package
- Count occurrences of unique values
- Check for duplicate values
- Build and install vignettes
- String filtering
- String filtering when using a database and dbplyr
- Create permutations of items from multple vectors
- Get class names of items in a dataframe/vector
- Printing more than default rows/columns from a table
- Search all objects, including functions, in global environment for string
- Search key words or phrases in help pages, vignettes or task views
- Update packages after updating R
- Timezone stuff
- Calculate minutes between two times
- filter for records within last n years
- View data exported by a package
- Browse vignettes for a given package
- Open function documentation from w/in RStudio
- Create a dataframe with random data
- Details about built-in data sets
- Use Github to search for packages using a particular function
- Conditional mutate
- extracting nested list into a tibble
- Scatterplot of all pairs
- Interactively explore plots
- Sort bar plot by counts
- Sort bar plot by counts - when using stat = “identity”
- Sort bar plots by counts, within facets, when using stat - “identity”
- Color coded correlation table
- filter correlations at a cutoff value
- plot distribution of all variables
- RMarkdown: insert date document was knitted
- Plot percentage of attributes that are NA for each outcome
- Plot the pecentage of rows that has at least 1 NA attribute, by outcome
- Plot the attributes (predictors) that are most likely to be missing
- Plot the attributes (predictors) that are most likely to be missing, by outcome
- Create a matrix that shows whether or not a particular combination of values is in the data
- Convert unix style epoch time to human readable time
- Clean Data: remove columns where no rows contain a value
- Print one plot for each data frame in a list column
- One plot for each attribute in a data frame - different scale for each attribute
- Get detailed information for each column in a database table
- Get just the columns and datatypes for each column in a database table
- Get just the column names
- Table of contents for rmarkdown document
- Get unique values from all non-numeric columns
- Get count and proportion of all non-numeric columns
- Get number of unique values for numeric columns
PDF of many of Rstudio’s cheat sheets
Another R cheat sheet I found useful
Selecting attributes from a data frame
# data [row, attribute_name]
iris[ 1, "Species"]
#> [1] setosa
#> Levels: setosa versicolor virginica
# approach 1: use [[ form of extract operator to extract a column
iris[["Species"]] %>%
head()
#> [1] setosa setosa setosa setosa setosa setosa
#> Levels: setosa versicolor virginica
# approach 2: use variable name in column dimension of data frame
iris[,"Species"] %>%
head()
#> [1] setosa setosa setosa setosa setosa setosa
#> Levels: setosa versicolor virginica
# approach 3: use the $ form of extract operator. Note that since this
iris$Species %>%
head()
#> [1] setosa setosa setosa setosa setosa setosa
#> Levels: setosa versicolor virginica
#Use individual variables in a pipe
ggplot2::diamonds %>%
.$cut %>%
head()
#> [1] Ideal Premium Good Premium Good Very Good
#> Levels: Fair < Good < Very Good < Premium < Ideal
ggplot2::diamonds %>%
dplyr::pull(cut) %>%
head()
#> [1] Ideal Premium Good Premium Good Very Good
#> Levels: Fair < Good < Very Good < Premium < Idealsubsetting rows
# approach 1: use exact row references
# dataframe[rows, columns]
mtcars[20:22,]
#> mpg cyl disp hp drat wt qsec vs am gear carb
#> Toyota Corolla 33.9 4 71.1 65 4.22 1.835 19.90 1 1 4 1
#> Toyota Corona 21.5 4 120.1 97 3.70 2.465 20.01 1 0 3 1
#> Dodge Challenger 15.5 8 318.0 150 2.76 3.520 16.87 0 0 3 2
# approach 2: use logic in the row dimension of reference
head(mtcars[mtcars$cyl == 4 & mtcars$am == 1,])
#> mpg cyl disp hp drat wt qsec vs am gear carb
#> Datsun 710 22.8 4 108.0 93 3.85 2.320 18.61 1 1 4 1
#> Fiat 128 32.4 4 78.7 66 4.08 2.200 19.47 1 1 4 1
#> Honda Civic 30.4 4 75.7 52 4.93 1.615 18.52 1 1 4 2
#> Toyota Corolla 33.9 4 71.1 65 4.22 1.835 19.90 1 1 4 1
#> Fiat X1-9 27.3 4 79.0 66 4.08 1.935 18.90 1 1 4 1
#> Porsche 914-2 26.0 4 120.3 91 4.43 2.140 16.70 0 1 5 2
head(mtcars[mtcars[,"cyl"] == 4,])
#> mpg cyl disp hp drat wt qsec vs am gear carb
#> Datsun 710 22.8 4 108.0 93 3.85 2.320 18.61 1 1 4 1
#> Merc 240D 24.4 4 146.7 62 3.69 3.190 20.00 1 0 4 2
#> Merc 230 22.8 4 140.8 95 3.92 3.150 22.90 1 0 4 2
#> Fiat 128 32.4 4 78.7 66 4.08 2.200 19.47 1 1 4 1
#> Honda Civic 30.4 4 75.7 52 4.93 1.615 18.52 1 1 4 2
#> Toyota Corolla 33.9 4 71.1 65 4.22 1.835 19.90 1 1 4 1
# approach 3: use which() function
theSubsetRows <- which(mtcars$cyl == 4)
head(mtcars[theSubsetRows,])
#> mpg cyl disp hp drat wt qsec vs am gear carb
#> Datsun 710 22.8 4 108.0 93 3.85 2.320 18.61 1 1 4 1
#> Merc 240D 24.4 4 146.7 62 3.69 3.190 20.00 1 0 4 2
#> Merc 230 22.8 4 140.8 95 3.92 3.150 22.90 1 0 4 2
#> Fiat 128 32.4 4 78.7 66 4.08 2.200 19.47 1 1 4 1
#> Honda Civic 30.4 4 75.7 52 4.93 1.615 18.52 1 1 4 2
#> Toyota Corolla 33.9 4 71.1 65 4.22 1.835 19.90 1 1 4 1
# approach 4: use output from a function that returns a logical
# array instead of row numbers as in the prior example
head(mtcars[!is.na(mtcars[,"cyl"]),])
#> mpg cyl disp hp drat wt qsec vs am gear carb
#> Mazda RX4 21.0 6 160 110 3.90 2.620 16.46 0 1 4 4
#> Mazda RX4 Wag 21.0 6 160 110 3.90 2.875 17.02 0 1 4 4
#> Datsun 710 22.8 4 108 93 3.85 2.320 18.61 1 1 4 1
#> Hornet 4 Drive 21.4 6 258 110 3.08 3.215 19.44 1 0 3 1
#> Hornet Sportabout 18.7 8 360 175 3.15 3.440 17.02 0 0 3 2
#> Valiant 18.1 6 225 105 2.76 3.460 20.22 1 0 3 1
# subset all rows with select columns
# dataframe[, c("column1", "column2")]
head(mtcars[, c("mpg", "carb")])
#> mpg carb
#> Mazda RX4 21.0 4
#> Mazda RX4 Wag 21.0 4
#> Datsun 710 22.8 1
#> Hornet 4 Drive 21.4 1
#> Hornet Sportabout 18.7 2
#> Valiant 18.1 1
# subset all rows where attribute matches
# dataframe[dataframe$attribute =="value", ]
mtcars[mtcars$cyl==4,]
#> mpg cyl disp hp drat wt qsec vs am gear carb
#> Datsun 710 22.8 4 108.0 93 3.85 2.320 18.61 1 1 4 1
#> Merc 240D 24.4 4 146.7 62 3.69 3.190 20.00 1 0 4 2
#> Merc 230 22.8 4 140.8 95 3.92 3.150 22.90 1 0 4 2
#> Fiat 128 32.4 4 78.7 66 4.08 2.200 19.47 1 1 4 1
#> Honda Civic 30.4 4 75.7 52 4.93 1.615 18.52 1 1 4 2
#> Toyota Corolla 33.9 4 71.1 65 4.22 1.835 19.90 1 1 4 1
#> Toyota Corona 21.5 4 120.1 97 3.70 2.465 20.01 1 0 3 1
#> Fiat X1-9 27.3 4 79.0 66 4.08 1.935 18.90 1 1 4 1
#> Porsche 914-2 26.0 4 120.3 91 4.43 2.140 16.70 0 1 5 2
#> Lotus Europa 30.4 4 95.1 113 3.77 1.513 16.90 1 1 5 2
#> Volvo 142E 21.4 4 121.0 109 4.11 2.780 18.60 1 1 4 2
# select all rows and all columns except for a few others
# in this example remove mpg and disp, the first and third columns
mtcars[,-c(1,3)]
#> cyl hp drat wt qsec vs am gear carb
#> Mazda RX4 6 110 3.90 2.620 16.46 0 1 4 4
#> Mazda RX4 Wag 6 110 3.90 2.875 17.02 0 1 4 4
#> Datsun 710 4 93 3.85 2.320 18.61 1 1 4 1
#> Hornet 4 Drive 6 110 3.08 3.215 19.44 1 0 3 1
#> Hornet Sportabout 8 175 3.15 3.440 17.02 0 0 3 2
#> Valiant 6 105 2.76 3.460 20.22 1 0 3 1
#> Duster 360 8 245 3.21 3.570 15.84 0 0 3 4
#> Merc 240D 4 62 3.69 3.190 20.00 1 0 4 2
#> Merc 230 4 95 3.92 3.150 22.90 1 0 4 2
#> Merc 280 6 123 3.92 3.440 18.30 1 0 4 4
#> Merc 280C 6 123 3.92 3.440 18.90 1 0 4 4
#> Merc 450SE 8 180 3.07 4.070 17.40 0 0 3 3
#> Merc 450SL 8 180 3.07 3.730 17.60 0 0 3 3
#> Merc 450SLC 8 180 3.07 3.780 18.00 0 0 3 3
#> Cadillac Fleetwood 8 205 2.93 5.250 17.98 0 0 3 4
#> Lincoln Continental 8 215 3.00 5.424 17.82 0 0 3 4
#> Chrysler Imperial 8 230 3.23 5.345 17.42 0 0 3 4
#> Fiat 128 4 66 4.08 2.200 19.47 1 1 4 1
#> Honda Civic 4 52 4.93 1.615 18.52 1 1 4 2
#> Toyota Corolla 4 65 4.22 1.835 19.90 1 1 4 1
#> Toyota Corona 4 97 3.70 2.465 20.01 1 0 3 1
#> Dodge Challenger 8 150 2.76 3.520 16.87 0 0 3 2
#> AMC Javelin 8 150 3.15 3.435 17.30 0 0 3 2
#> Camaro Z28 8 245 3.73 3.840 15.41 0 0 3 4
#> Pontiac Firebird 8 175 3.08 3.845 17.05 0 0 3 2
#> Fiat X1-9 4 66 4.08 1.935 18.90 1 1 4 1
#> Porsche 914-2 4 91 4.43 2.140 16.70 0 1 5 2
#> Lotus Europa 4 113 3.77 1.513 16.90 1 1 5 2
#> Ford Pantera L 8 264 4.22 3.170 14.50 0 1 5 4
#> Ferrari Dino 6 175 3.62 2.770 15.50 0 1 5 6
#> Maserati Bora 8 335 3.54 3.570 14.60 0 1 5 8
#> Volvo 142E 4 109 4.11 2.780 18.60 1 1 4 2
#alternate - if all rows you want to remove are next to each other
mtcars[,-(1:3)]
#> hp drat wt qsec vs am gear carb
#> Mazda RX4 110 3.90 2.620 16.46 0 1 4 4
#> Mazda RX4 Wag 110 3.90 2.875 17.02 0 1 4 4
#> Datsun 710 93 3.85 2.320 18.61 1 1 4 1
#> Hornet 4 Drive 110 3.08 3.215 19.44 1 0 3 1
#> Hornet Sportabout 175 3.15 3.440 17.02 0 0 3 2
#> Valiant 105 2.76 3.460 20.22 1 0 3 1
#> Duster 360 245 3.21 3.570 15.84 0 0 3 4
#> Merc 240D 62 3.69 3.190 20.00 1 0 4 2
#> Merc 230 95 3.92 3.150 22.90 1 0 4 2
#> Merc 280 123 3.92 3.440 18.30 1 0 4 4
#> Merc 280C 123 3.92 3.440 18.90 1 0 4 4
#> Merc 450SE 180 3.07 4.070 17.40 0 0 3 3
#> Merc 450SL 180 3.07 3.730 17.60 0 0 3 3
#> Merc 450SLC 180 3.07 3.780 18.00 0 0 3 3
#> Cadillac Fleetwood 205 2.93 5.250 17.98 0 0 3 4
#> Lincoln Continental 215 3.00 5.424 17.82 0 0 3 4
#> Chrysler Imperial 230 3.23 5.345 17.42 0 0 3 4
#> Fiat 128 66 4.08 2.200 19.47 1 1 4 1
#> Honda Civic 52 4.93 1.615 18.52 1 1 4 2
#> Toyota Corolla 65 4.22 1.835 19.90 1 1 4 1
#> Toyota Corona 97 3.70 2.465 20.01 1 0 3 1
#> Dodge Challenger 150 2.76 3.520 16.87 0 0 3 2
#> AMC Javelin 150 3.15 3.435 17.30 0 0 3 2
#> Camaro Z28 245 3.73 3.840 15.41 0 0 3 4
#> Pontiac Firebird 175 3.08 3.845 17.05 0 0 3 2
#> Fiat X1-9 66 4.08 1.935 18.90 1 1 4 1
#> Porsche 914-2 91 4.43 2.140 16.70 0 1 5 2
#> Lotus Europa 113 3.77 1.513 16.90 1 1 5 2
#> Ford Pantera L 264 4.22 3.170 14.50 0 1 5 4
#> Ferrari Dino 175 3.62 2.770 15.50 0 1 5 6
#> Maserati Bora 335 3.54 3.570 14.60 0 1 5 8
#> Volvo 142E 109 4.11 2.780 18.60 1 1 4 2Filtering / extracting / subsetting data frames based on attribute value
mtcars[which(mtcars$cyl == 4), "mpg"]
#> [1] 22.8 24.4 22.8 32.4 30.4 33.9 21.5 27.3 26.0 30.4 21.4
subset(mtcars$mpg, mtcars$cyl == 4)
#> [1] 22.8 24.4 22.8 32.4 30.4 33.9 21.5 27.3 26.0 30.4 21.4Filtering for rows where at least one column is missing
dplyr::starwars %>%
dplyr::filter(dplyr::if_any(dplyr::everything(), ~is.na(.x)))
#> # A tibble: 58 x 14
#> name height mass hair_color skin_color eye_color birth_year sex gender
#> <chr> <int> <dbl> <chr> <chr> <chr> <dbl> <chr> <chr>
#> 1 C-3PO 167 75 <NA> gold yellow 112 none mascu~
#> 2 R2-D2 96 32 <NA> white, bl~ red 33 none mascu~
#> 3 R5-D4 97 32 <NA> white, red red NA none mascu~
#> 4 Wilhuf~ 180 NA auburn, gr~ fair blue 64 male mascu~
#> 5 Greedo 173 74 <NA> green black 44 male mascu~
#> 6 Jabba ~ 175 1358 <NA> green-tan~ orange 600 herm~ mascu~
#> 7 Jek To~ 180 110 brown fair blue NA male mascu~
#> 8 Yoda 66 17 white green brown 896 male mascu~
#> 9 IG-88 200 140 none metal red 15 none mascu~
#> 10 Mon Mo~ 150 NA auburn fair blue 48 fema~ femin~
#> # ... with 48 more rows, and 5 more variables: homeworld <chr>, species <chr>,
#> # films <list>, vehicles <list>, starships <list>Filtering for rows where all columns have values
dplyr::starwars %>%
tidyr::drop_na()
#> # A tibble: 6 x 14
#> name height mass hair_color skin_color eye_color birth_year sex gender
#> <chr> <int> <dbl> <chr> <chr> <chr> <dbl> <chr> <chr>
#> 1 Luke Sk~ 172 77 blond fair blue 19 male mascu~
#> 2 Obi-Wan~ 182 77 auburn, wh~ fair blue-gray 57 male mascu~
#> 3 Anakin ~ 188 84 blond fair blue 41.9 male mascu~
#> 4 Chewbac~ 228 112 brown unknown blue 200 male mascu~
#> 5 Wedge A~ 170 77 brown fair hazel 21 male mascu~
#> 6 Darth M~ 175 80 none red yellow 54 male mascu~
#> # ... with 5 more variables: homeworld <chr>, species <chr>, films <list>,
#> # vehicles <list>, starships <list>Exploring data
#summary stats from pastecs package
pastecs::stat.desc(mtcars)
#> mpg cyl disp hp drat
#> nbr.val 32.0000000 32.0000000 3.200000e+01 32.0000000 32.00000000
#> nbr.null 0.0000000 0.0000000 0.000000e+00 0.0000000 0.00000000
#> nbr.na 0.0000000 0.0000000 0.000000e+00 0.0000000 0.00000000
#> min 10.4000000 4.0000000 7.110000e+01 52.0000000 2.76000000
#> max 33.9000000 8.0000000 4.720000e+02 335.0000000 4.93000000
#> range 23.5000000 4.0000000 4.009000e+02 283.0000000 2.17000000
#> sum 642.9000000 198.0000000 7.383100e+03 4694.0000000 115.09000000
#> median 19.2000000 6.0000000 1.963000e+02 123.0000000 3.69500000
#> mean 20.0906250 6.1875000 2.307219e+02 146.6875000 3.59656250
#> SE.mean 1.0654240 0.3157093 2.190947e+01 12.1203173 0.09451874
#> CI.mean.0.95 2.1729465 0.6438934 4.468466e+01 24.7195501 0.19277224
#> var 36.3241028 3.1895161 1.536080e+04 4700.8669355 0.28588135
#> std.dev 6.0269481 1.7859216 1.239387e+02 68.5628685 0.53467874
#> coef.var 0.2999881 0.2886338 5.371779e-01 0.4674077 0.14866382
#> wt qsec vs am gear
#> nbr.val 32.0000000 32.0000000 32.00000000 32.00000000 32.0000000
#> nbr.null 0.0000000 0.0000000 18.00000000 19.00000000 0.0000000
#> nbr.na 0.0000000 0.0000000 0.00000000 0.00000000 0.0000000
#> min 1.5130000 14.5000000 0.00000000 0.00000000 3.0000000
#> max 5.4240000 22.9000000 1.00000000 1.00000000 5.0000000
#> range 3.9110000 8.4000000 1.00000000 1.00000000 2.0000000
#> sum 102.9520000 571.1600000 14.00000000 13.00000000 118.0000000
#> median 3.3250000 17.7100000 0.00000000 0.00000000 4.0000000
#> mean 3.2172500 17.8487500 0.43750000 0.40625000 3.6875000
#> SE.mean 0.1729685 0.3158899 0.08909831 0.08820997 0.1304266
#> CI.mean.0.95 0.3527715 0.6442617 0.18171719 0.17990541 0.2660067
#> var 0.9573790 3.1931661 0.25403226 0.24899194 0.5443548
#> std.dev 0.9784574 1.7869432 0.50401613 0.49899092 0.7378041
#> coef.var 0.3041285 0.1001159 1.15203687 1.22828533 0.2000825
#> carb
#> nbr.val 32.0000000
#> nbr.null 0.0000000
#> nbr.na 0.0000000
#> min 1.0000000
#> max 8.0000000
#> range 7.0000000
#> sum 90.0000000
#> median 2.0000000
#> mean 2.8125000
#> SE.mean 0.2855297
#> CI.mean.0.95 0.5823417
#> var 2.6088710
#> std.dev 1.6152000
#> coef.var 0.5742933
#Counting NULL values in column
sum(is.na(mtcars$cyl))
#> [1] 0Prevent garbage characters when using read.csv on data exported from SQL
#SOURCE: http://stackoverflow.com/questions/24568056/rs-read-csv-prepending-1st-column-name-with-junk-text
read.csv(file = "my_file.csv", fileEncoding = "UTF-8-BOM")List functions exported by a loaded package
head(ls("package:base"))
#> [1] "-" "-.Date" "-.POSIXt" "!" "!.hexmode" "!.octmode"Count occurrences of unique values
table(mtcars$cyl)
#>
#> 4 6 8
#> 11 7 14
dplyr::count(mtcars, cyl)
#> cyl n
#> 1 4 11
#> 2 6 7
#> 3 8 14
as.data.frame(table(mtcars$cyl))
#> Var1 Freq
#> 1 4 11
#> 2 6 7
#> 3 8 14
dplyr::count(mtcars, cyl, gear)
#> cyl gear n
#> 1 4 3 1
#> 2 4 4 8
#> 3 4 5 2
#> 4 6 3 2
#> 5 6 4 4
#> 6 6 5 1
#> 7 8 3 12
#> 8 8 5 2
xtabs(~cyl + gear, mtcars)
#> gear
#> cyl 3 4 5
#> 4 1 8 2
#> 6 2 4 1
#> 8 12 0 2
#Get table with row and column sums
addmargins(table(mtcars$cyl,useNA = "always"))
#>
#> 4 6 8 <NA> Sum
#> 11 7 14 0 32
#Get percentage table
addmargins(sort(prop.table(table(mtcars$cyl,useNA = "always"))))
#>
#> <NA> 6 4 8 Sum
#> 0.00000 0.21875 0.34375 0.43750 1.00000Check for duplicate values
length(unique(nps$email))==nrow(nps)
#pull duplicate values
nycflights13::planes %>%
dplyr::count(tailnum) %>%
dplyr::filter(n > 1)
nycflights13::weather %>%
dplyr::count(year, month, day, hour, origin) %>%
filter(n > 1)Build and install vignettes
devtools::install(build_vignettes = TRUE)String filtering
#filter output of names - or any other character vector
row.names(mtcars)[grepl("Merc",row.names(mtcars))]
#> [1] "Merc 240D" "Merc 230" "Merc 280" "Merc 280C" "Merc 450SE"
#> [6] "Merc 450SL" "Merc 450SLC"
names(mtcars)[grepl("c", names(mtcars))]
#> [1] "cyl" "qsec" "carb"
grep("C", names(mtcars), ignore.case = TRUE, value = TRUE)
#> [1] "cyl" "qsec" "carb"
grep("C", names(mtcars), ignore.case = TRUE, value = FALSE)
#> [1] 2 7 11
#Filter with dplyr where row cointains string
dplyr::filter(ggplot2::diamonds, grepl('Good', cut))
#> # A tibble: 16,988 x 10
#> carat cut color clarity depth table price x y z
#> <dbl> <ord> <ord> <ord> <dbl> <dbl> <int> <dbl> <dbl> <dbl>
#> 1 0.23 Good E VS1 56.9 65 327 4.05 4.07 2.31
#> 2 0.31 Good J SI2 63.3 58 335 4.34 4.35 2.75
#> 3 0.24 Very Good J VVS2 62.8 57 336 3.94 3.96 2.48
#> 4 0.24 Very Good I VVS1 62.3 57 336 3.95 3.98 2.47
#> 5 0.26 Very Good H SI1 61.9 55 337 4.07 4.11 2.53
#> 6 0.23 Very Good H VS1 59.4 61 338 4 4.05 2.39
#> 7 0.3 Good J SI1 64 55 339 4.25 4.28 2.73
#> 8 0.3 Good J SI1 63.4 54 351 4.23 4.29 2.7
#> 9 0.3 Good J SI1 63.8 56 351 4.23 4.26 2.71
#> 10 0.3 Very Good J SI1 62.7 59 351 4.21 4.27 2.66
#> # ... with 16,978 more rows
#Filter column/attribute of data frame for regex text pattern match
nycflights13::airports %>%
dplyr::filter(stringr::str_detect(name, "County")) %>%
head()
#> # A tibble: 6 x 8
#> faa name lat lon alt tz dst tzone
#> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <chr> <chr>
#> 1 0G6 Williams County Airport 41.5 -84.5 730 -5 A America/New_Y~
#> 2 0S9 Jefferson County Intl 48.1 -123. 108 -8 A America/Los_A~
#> 3 0W3 Harford County Airport 39.6 -76.2 409 -5 A America/New_Y~
#> 4 17G Port Bucyrus-Crawford Cou~ 40.8 -83.0 1003 -5 A America/New_Y~
#> 5 19A Jackson County Airport 34.2 -83.6 951 -5 U America/New_Y~
#> 6 24J Suwannee County Airport 30.3 -83.0 104 -5 A America/New_Y~
#Search for, and extract matches
pattern <- "[[:upper:]]"
stringr::sentences %>%
stringr::str_subset(pattern) %>%
stringr::str_extract_all(pattern) %>%
head()
#> [[1]]
#> [1] "T"
#>
#> [[2]]
#> [1] "G"
#>
#> [[3]]
#> [1] "I"
#>
#> [[4]]
#> [1] "T"
#>
#> [[5]]
#> [1] "R"
#>
#> [[6]]
#> [1] "T"
#filter column/attriubte names based on string
ggplot2::diamonds %>%
dplyr::select_at(dplyr::vars(dplyr::contains('c')))
#> # A tibble: 53,940 x 5
#> carat cut color clarity price
#> <dbl> <ord> <ord> <ord> <int>
#> 1 0.23 Ideal E SI2 326
#> 2 0.21 Premium E SI1 326
#> 3 0.23 Good E VS1 327
#> 4 0.29 Premium I VS2 334
#> 5 0.31 Good J SI2 335
#> 6 0.24 Very Good J VVS2 336
#> 7 0.24 Very Good I VVS1 336
#> 8 0.26 Very Good H SI1 337
#> 9 0.22 Fair E VS2 337
#> 10 0.23 Very Good H VS1 338
#> # ... with 53,930 more rows
#filter where attribute value starts with a string
mtcars %>%
tibble::rownames_to_column("model") %>%
dplyr::filter(stringr::str_detect(model, "^Merc"))
#> model mpg cyl disp hp drat wt qsec vs am gear carb
#> 1 Merc 240D 24.4 4 146.7 62 3.69 3.19 20.0 1 0 4 2
#> 2 Merc 230 22.8 4 140.8 95 3.92 3.15 22.9 1 0 4 2
#> 3 Merc 280 19.2 6 167.6 123 3.92 3.44 18.3 1 0 4 4
#> 4 Merc 280C 17.8 6 167.6 123 3.92 3.44 18.9 1 0 4 4
#> 5 Merc 450SE 16.4 8 275.8 180 3.07 4.07 17.4 0 0 3 3
#> 6 Merc 450SL 17.3 8 275.8 180 3.07 3.73 17.6 0 0 3 3
#> 7 Merc 450SLC 15.2 8 275.8 180 3.07 3.78 18.0 0 0 3 3
#rename columns/attributes if they contain certain pattern
#in this case replace '.' with '_'
#SOU
nycflights13::weather %>%
dplyr::rename_at(
dplyr::vars(dplyr::contains('_')), dplyr::funs(stringr::str_replace(., "_", "\\."))
) %>%
names()
#> Warning: `funs()` was deprecated in dplyr 0.8.0.
#> Please use a list of either functions or lambdas:
#>
#> # Simple named list:
#> list(mean = mean, median = median)
#>
#> # Auto named with `tibble::lst()`:
#> tibble::lst(mean, median)
#>
#> # Using lambdas
#> list(~ mean(., trim = .2), ~ median(., na.rm = TRUE))
#> [1] "origin" "year" "month" "day" "hour"
#> [6] "temp" "dewp" "humid" "wind.dir" "wind.speed"
#> [11] "wind.gust" "precip" "pressure" "visib" "time.hour"String filtering when using a database and dbplyr
The previous mentions won’t always work when filtering against data in a database. Reference: https://github.com/tidyverse/dplyr/issues/3090
Solution from: https://stackoverflow.com/questions/38962585/pass-sql-functions-in-dplyr-filter-function-on-database/47198795#47198795
#load some data into a temp database
con <- DBI::dbConnect(RSQLite::SQLite(), path = ":memory:")
DBI::dbWriteTable(con, "airports", nycflights13::airports)
#query the data - name contains "Island" - this is case sensitive
dplyr::tbl(con, "airports") %>%
dplyr::filter(name %like% "%Island%") %>%
head(5)
#> # Source: lazy query [?? x 8]
#> # Database: sqlite 3.35.5 []
#> faa name lat lon alt tz dst tzone
#> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <chr> <chr>
#> 1 09J Jekyll Island Airport 31.1 -81.4 11 -5 A America/New_~
#> 2 2B2 Plum Island Airport 42.8 -70.8 11 -5 A America/New_~
#> 3 ANN Annette Island 55.0 -132. 119 -9 A America/Anch~
#> 4 BID Block Island State Airport 41.2 -71.6 108 -5 A America/New_~
#> 5 BRO Brownsville South Padre Is~ 25.9 -97.4 22 -6 A America/Chic~This works when I test against a Microsoft SQL database but not SQLite. Returns error:
Error in stri_detect_regex(string, pattern, opts_regex = opts(pattern)) : object 'name' not found
#load some data into a temp database
con <- DBI::dbConnect(RSQLite::SQLite(), path = ":memory:")
DBI::dbWriteTable(con, "airports", nycflights13::airports)
#query the data - name contains "Island" - this is case sensitive
dplyr::tbl(con, "airports") %>%
dplyr::filter(stringr::str_detect(name, "Island")) %>%
head(5)Create permutations of items from multple vectors
expand.grid(
c(1, 2),
c(3, 4),
c(2, 3))
#> Var1 Var2 Var3
#> 1 1 3 2
#> 2 2 3 2
#> 3 1 4 2
#> 4 2 4 2
#> 5 1 3 3
#> 6 2 3 3
#> 7 1 4 3
#> 8 2 4 3Get class names of items in a dataframe/vector
sapply(mtcars, class)
#> mpg cyl disp hp drat wt qsec vs
#> "numeric" "numeric" "numeric" "numeric" "numeric" "numeric" "numeric" "numeric"
#> am gear carb
#> "numeric" "numeric" "numeric"Printing more than default rows/columns from a table
mtcars %>% dplyr::as_tibble() %>% print(n=15, width = Inf)
#> # A tibble: 32 x 11
#> mpg cyl disp hp drat wt qsec vs am gear carb
#> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 21 6 160 110 3.9 2.62 16.5 0 1 4 4
#> 2 21 6 160 110 3.9 2.88 17.0 0 1 4 4
#> 3 22.8 4 108 93 3.85 2.32 18.6 1 1 4 1
#> 4 21.4 6 258 110 3.08 3.22 19.4 1 0 3 1
#> 5 18.7 8 360 175 3.15 3.44 17.0 0 0 3 2
#> 6 18.1 6 225 105 2.76 3.46 20.2 1 0 3 1
#> 7 14.3 8 360 245 3.21 3.57 15.8 0 0 3 4
#> 8 24.4 4 147. 62 3.69 3.19 20 1 0 4 2
#> 9 22.8 4 141. 95 3.92 3.15 22.9 1 0 4 2
#> 10 19.2 6 168. 123 3.92 3.44 18.3 1 0 4 4
#> 11 17.8 6 168. 123 3.92 3.44 18.9 1 0 4 4
#> 12 16.4 8 276. 180 3.07 4.07 17.4 0 0 3 3
#> 13 17.3 8 276. 180 3.07 3.73 17.6 0 0 3 3
#> 14 15.2 8 276. 180 3.07 3.78 18 0 0 3 3
#> 15 10.4 8 472 205 2.93 5.25 18.0 0 0 3 4
#> # ... with 17 more rowsSearch all objects, including functions, in global environment for string
apropos("cars")
#> [1] "cars" "mtcars"Search key words or phrases in help pages, vignettes or task views
RSiteSearch("Microsoft AND SQL")Update packages after updating R
update.packages(ask = FALSE, checkBuilt = TRUE)Timezone stuff
#Get timezone
Sys.timezone()
#> [1] "America/New_York"
#Get all timezones
OlsonNames() %>% head()
#> [1] "Africa/Abidjan" "Africa/Accra" "Africa/Addis_Ababa"
#> [4] "Africa/Algiers" "Africa/Asmara" "Africa/Asmera"Calculate minutes between two times
library(lubridate)
#>
#> Attaching package: 'lubridate'
#> The following objects are masked from 'package:base':
#>
#> date, intersect, setdiff, union
ymd_hms("2018-10-25 12:00:00") %--% ymd_hms("2018-10-25 13:00:00") / dminutes(1)
#> [1] 60filter for records within last n years
works with dbplyr too this is not a great example but I wanted it to work for at least a few more years.
hundred_years_ago <- lubridate::today() - lubridate::years(100)
nycflights13::flights %>%
dplyr::filter(time_hour >= hundred_years_ago)
#> # A tibble: 336,776 x 19
#> year month day dep_time sched_dep_time dep_delay arr_time sched_arr_time
#> <int> <int> <int> <int> <int> <dbl> <int> <int>
#> 1 2013 1 1 517 515 2 830 819
#> 2 2013 1 1 533 529 4 850 830
#> 3 2013 1 1 542 540 2 923 850
#> 4 2013 1 1 544 545 -1 1004 1022
#> 5 2013 1 1 554 600 -6 812 837
#> 6 2013 1 1 554 558 -4 740 728
#> 7 2013 1 1 555 600 -5 913 854
#> 8 2013 1 1 557 600 -3 709 723
#> 9 2013 1 1 557 600 -3 838 846
#> 10 2013 1 1 558 600 -2 753 745
#> # ... with 336,766 more rows, and 11 more variables: arr_delay <dbl>,
#> # carrier <chr>, flight <int>, tailnum <chr>, origin <chr>, dest <chr>,
#> # air_time <dbl>, distance <dbl>, hour <dbl>, minute <dbl>, time_hour <dttm>View data exported by a package
data(package = "ggplot2")Browse vignettes for a given package
browseVignettes(package = "dplyr")Open function documentation from w/in RStudio
Use F2
Create a dataframe with random data
#the call to set.seed ensures anyone who runs the code will get the same random number stream.
set.seed(1)
data.frame(x = rnorm(4), y = rnorm(4), z = sample(LETTERS, 4))
#> x y z
#> 1 -0.6264538 0.3295078 V
#> 2 0.1836433 -0.8204684 N
#> 3 -0.8356286 0.4874291 J
#> 4 1.5952808 0.7383247 GDetails about built-in data sets
library(help = "datasets")
data()Use Github to search for packages using a particular function
Put this into the GitHub search box to see how packages on CRAN use the llply() function from plyr
"llply" user:cran language:RConditional mutate
NOTE: should probably also look at recode here
nycflights13::airlines %>%
dplyr::mutate(
name = dplyr::case_when(
name == "Virgin America" ~ "Alaska Airlines Inc.",
TRUE ~ name)
)
#> # A tibble: 16 x 2
#> carrier name
#> <chr> <chr>
#> 1 9E Endeavor Air Inc.
#> 2 AA American Airlines Inc.
#> 3 AS Alaska Airlines Inc.
#> 4 B6 JetBlue Airways
#> 5 DL Delta Air Lines Inc.
#> 6 EV ExpressJet Airlines Inc.
#> 7 F9 Frontier Airlines Inc.
#> 8 FL AirTran Airways Corporation
#> 9 HA Hawaiian Airlines Inc.
#> 10 MQ Envoy Air
#> 11 OO SkyWest Airlines Inc.
#> 12 UA United Air Lines Inc.
#> 13 US US Airways Inc.
#> 14 VX Alaska Airlines Inc.
#> 15 WN Southwest Airlines Co.
#> 16 YV Mesa Airlines Inc.extracting nested list into a tibble
SOURCE: https://cfss.uchicago.edu/webdata004_simplifying_lists.html
my_list <- list(
list(first = "nick", last = "vasile", weight = 170),
list(first = "bob", last = "smith", weight = 150)
)
my_list
#> [[1]]
#> [[1]]$first
#> [1] "nick"
#>
#> [[1]]$last
#> [1] "vasile"
#>
#> [[1]]$weight
#> [1] 170
#>
#>
#> [[2]]
#> [[2]]$first
#> [1] "bob"
#>
#> [[2]]$last
#> [1] "smith"
#>
#> [[2]]$weight
#> [1] 150
purrr::map_df(my_list, magrittr::extract)
#> # A tibble: 2 x 3
#> first last weight
#> <chr> <chr> <dbl>
#> 1 nick vasile 170
#> 2 bob smith 150Scatterplot of all pairs
pairs(mtcars)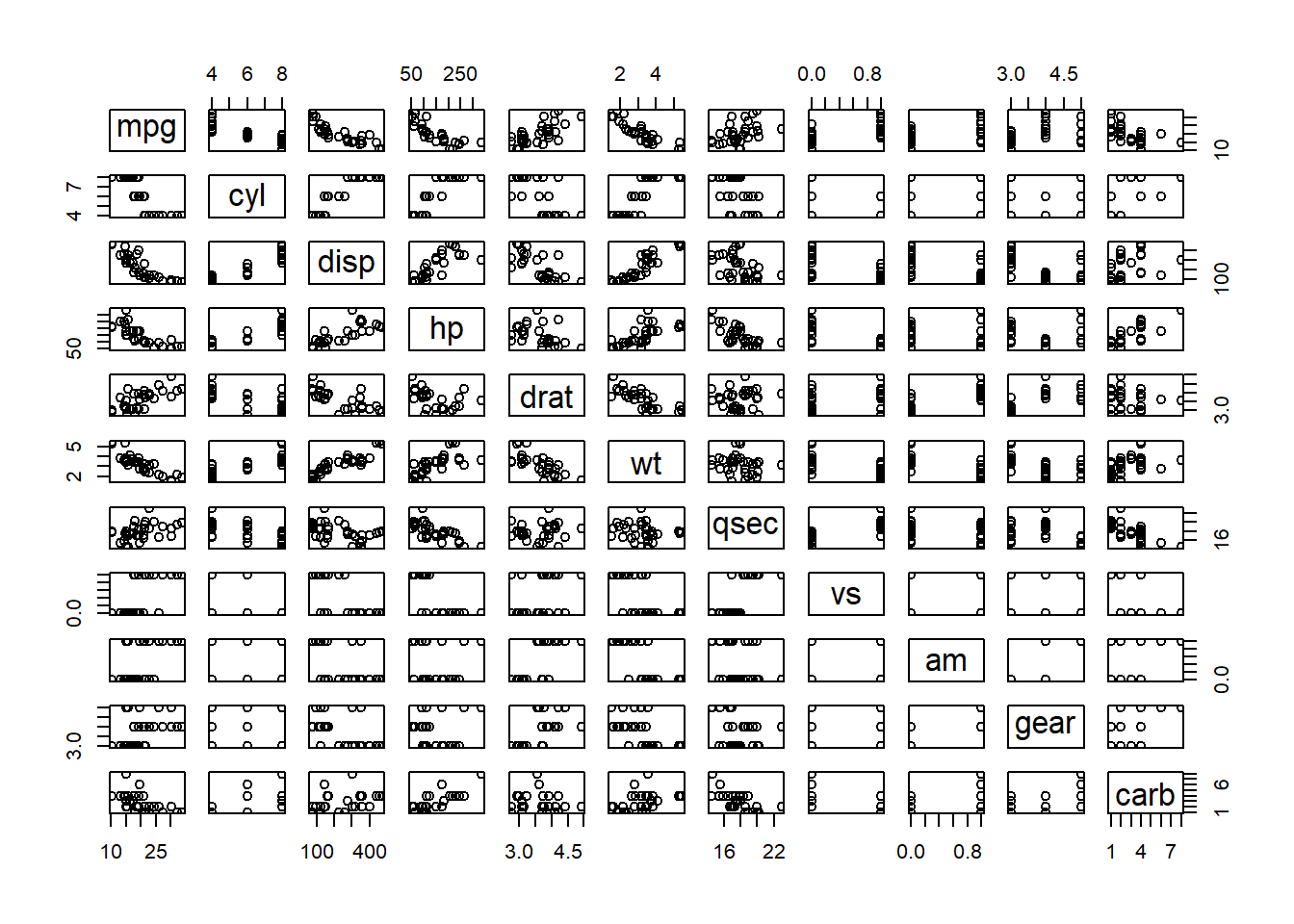
Interactively explore plots
identify(mtcars$hp, mtcars$mpg, mtcars$mpg)Sort bar plot by counts
nycflights13::flights %>%
ggplot2::ggplot(ggplot2::aes(x=forcats::fct_infreq(carrier))) +
ggplot2::geom_bar()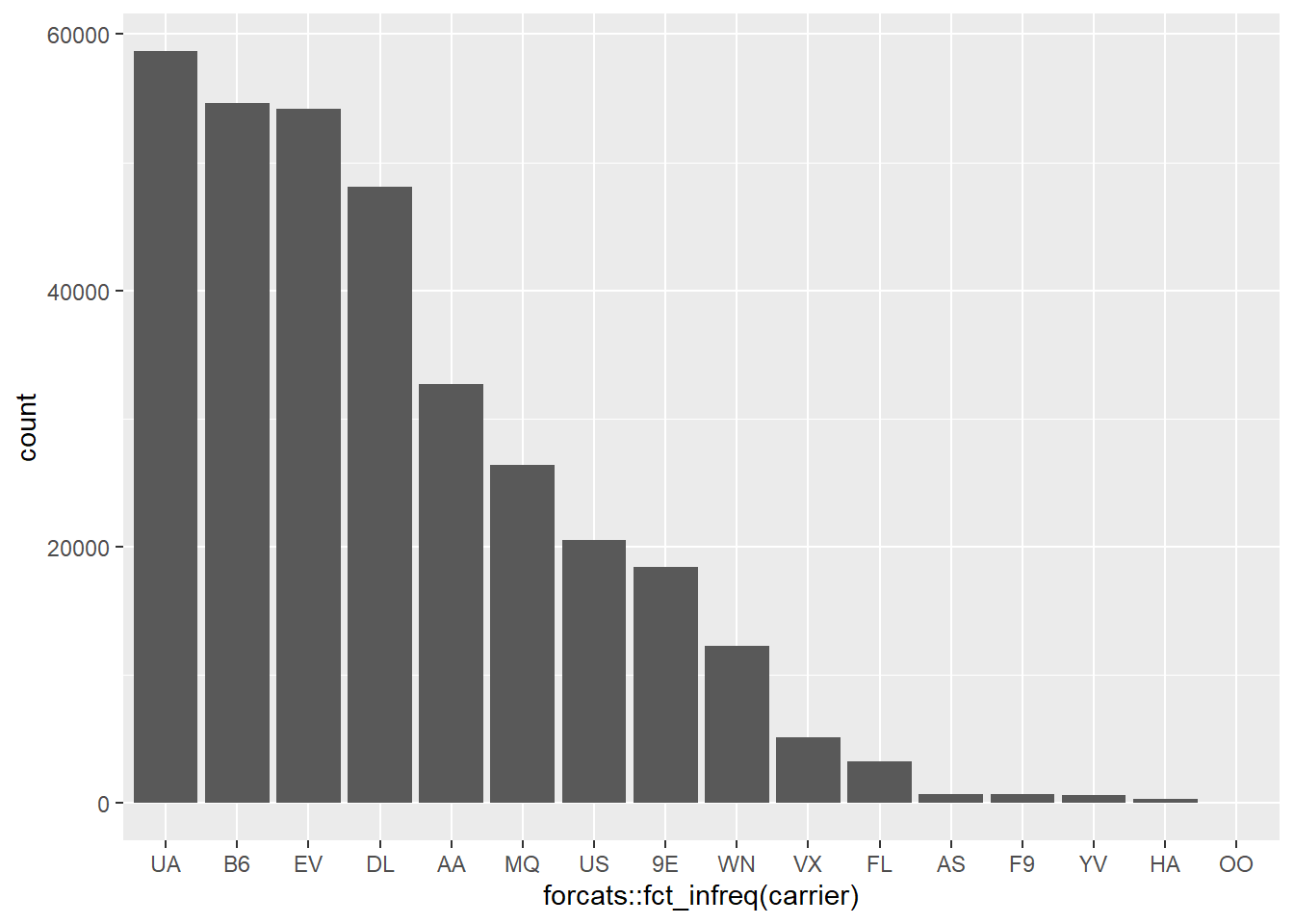
nycflights13::flights %>%
ggplot2::ggplot(ggplot2::aes(x=forcats::fct_rev(
forcats::fct_infreq(carrier)))) +
ggplot2::geom_bar() +
ggplot2::coord_flip()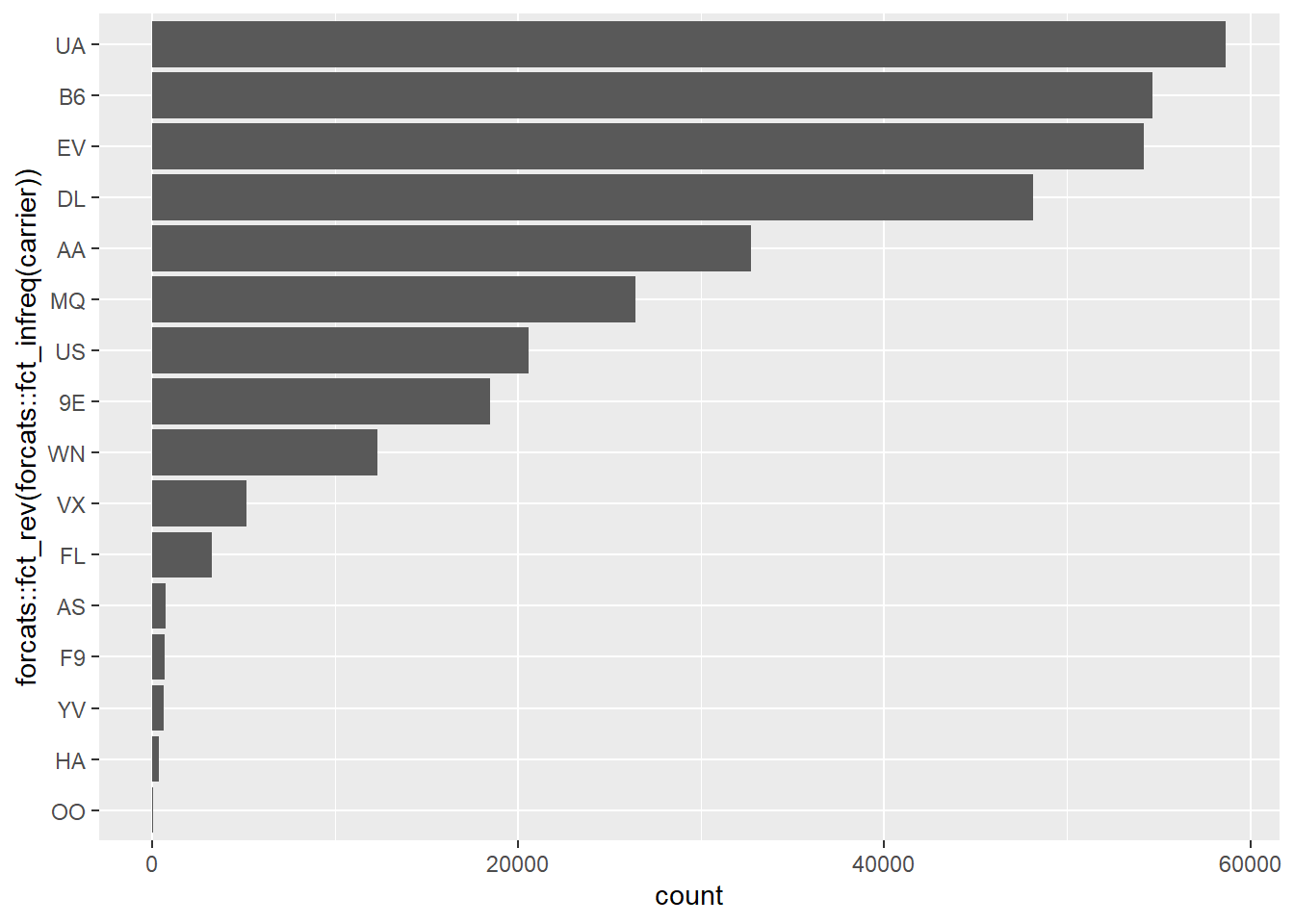
Sort bar plot by counts - when using stat = “identity”
avg_delay_by_carrier <- nycflights13::flights %>%
dplyr::group_by(carrier) %>%
dplyr::summarize(mean_delay = mean(dep_delay, na.rm = TRUE))
avg_delay_by_carrier %>%
ggplot2::ggplot(ggplot2::aes(x=reorder(carrier, -mean_delay),
y = mean_delay)) +
ggplot2::geom_bar(stat="identity") +
ggplot2::labs(x = "carrier") # if you want to set x-axis to original attribute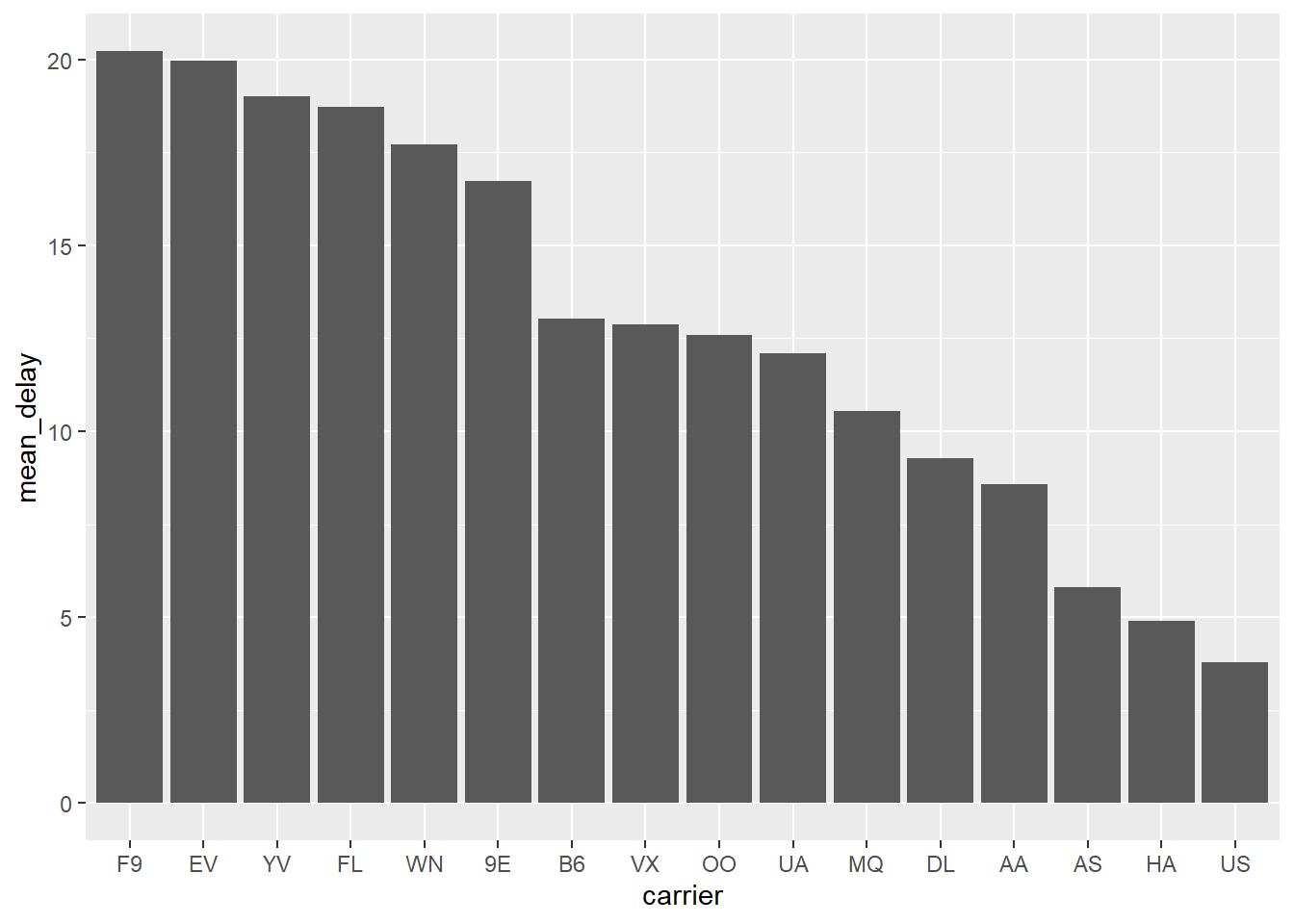
avg_delay_by_carrier <-
nycflights13::flights %>%
dplyr::group_by(carrier) %>%
dplyr::summarize(mean_delay = mean(dep_delay, na.rm = TRUE))
avg_delay_by_carrier %>%
ggplot2::ggplot(ggplot2::aes(x=reorder(carrier, mean_delay),
y = mean_delay)) +
ggplot2::geom_bar(stat="identity") +
ggplot2::coord_flip()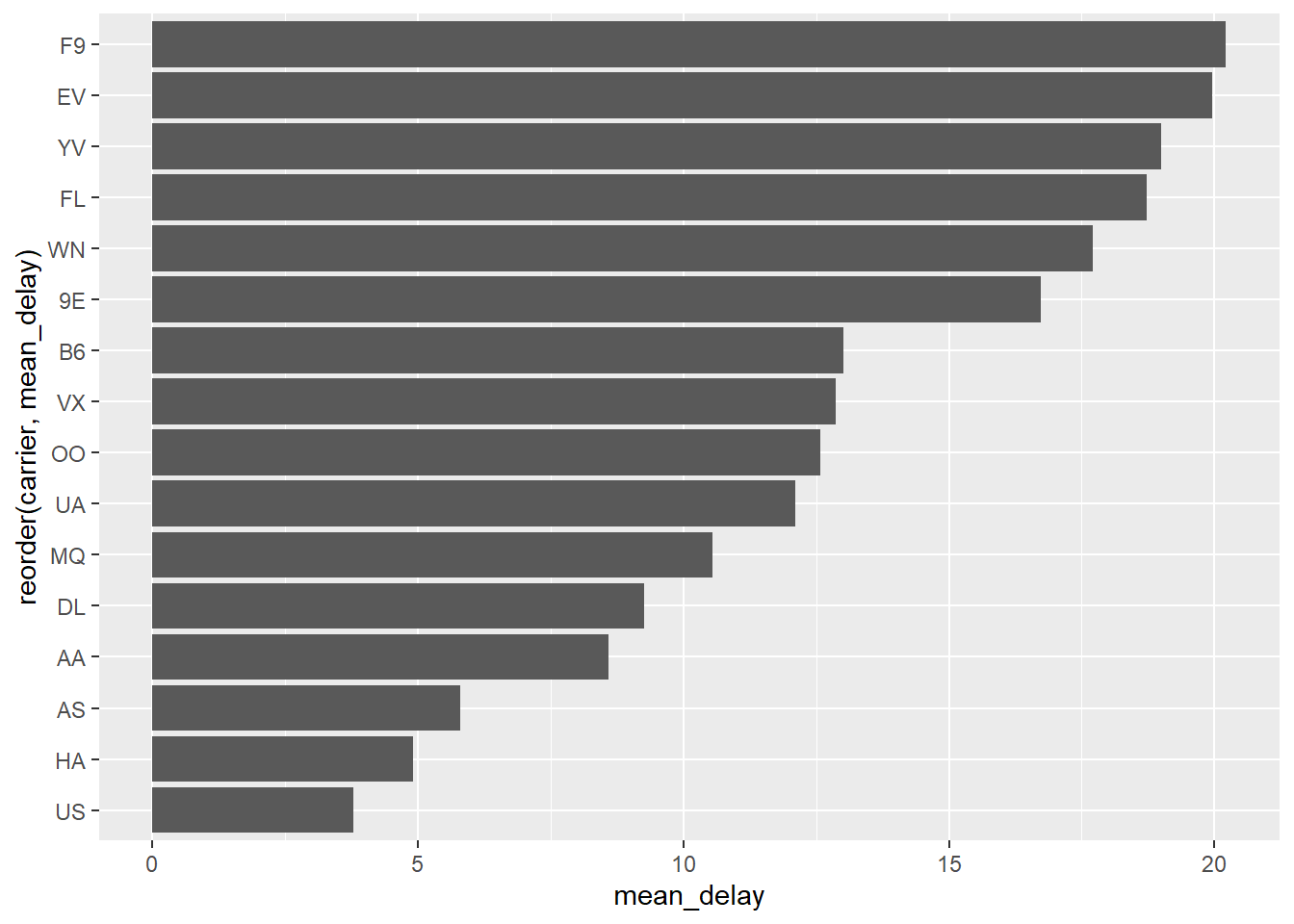
Sort bar plots by counts, within facets, when using stat - “identity”
SOURCE: https://www.programmingwithr.com/how-to-reorder-arrange-bars-with-in-each-facet-of-ggplot/
iris_gathered <-
tidyr::gather(iris, metric, value, -Species)
iris_gathered %>%
ggplot2::ggplot(ggplot2::aes(tidytext::reorder_within(Species, value, metric),
value)) +
ggplot2::geom_bar(stat = 'identity') +
tidytext::scale_x_reordered() +
ggplot2::coord_flip() +
ggplot2::facet_wrap(~ metric, scales = "free") +
ggplot2::xlab("species")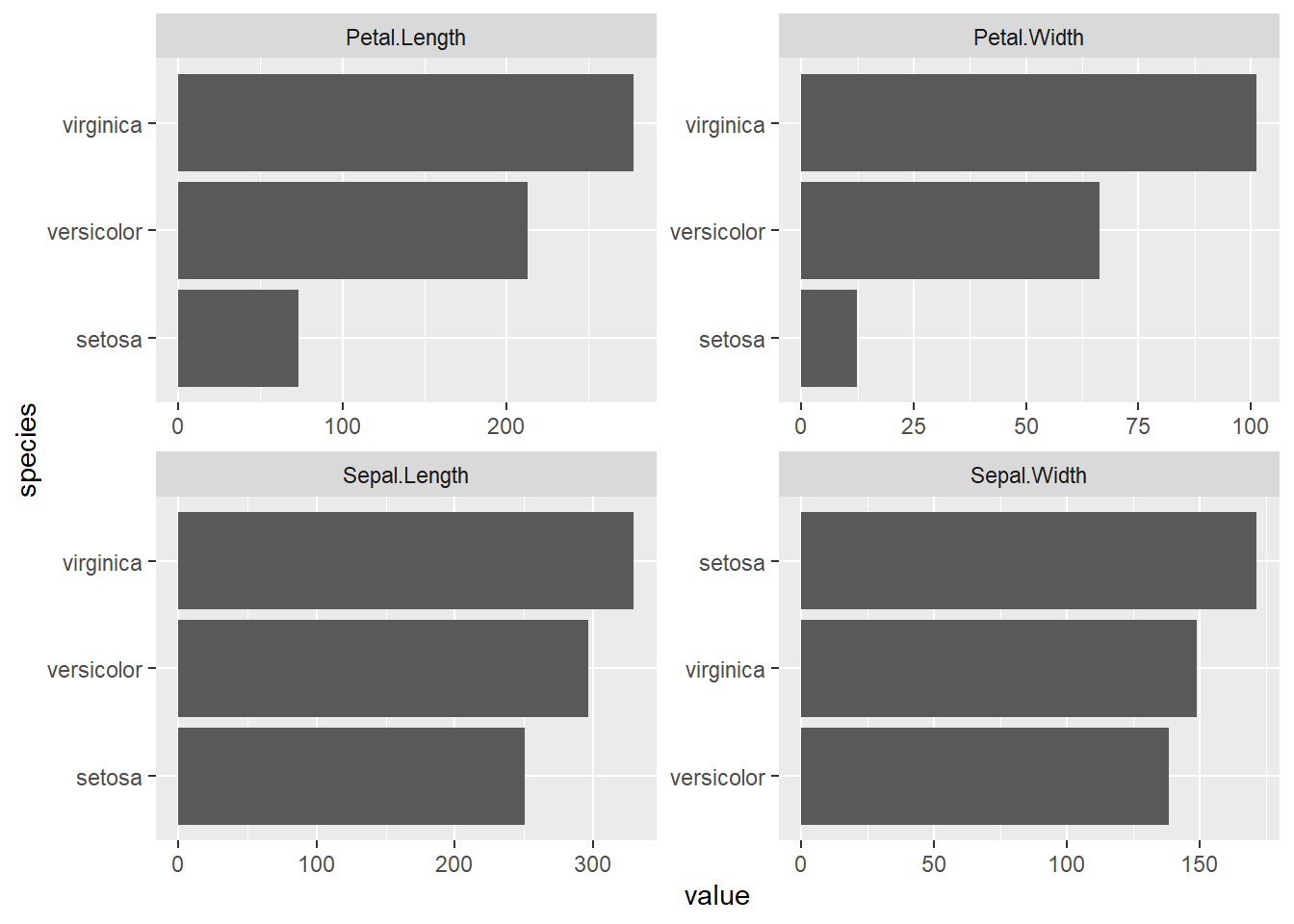
Color coded correlation table
M <- cor(mtcars)
corrplot::corrplot(M, method="color")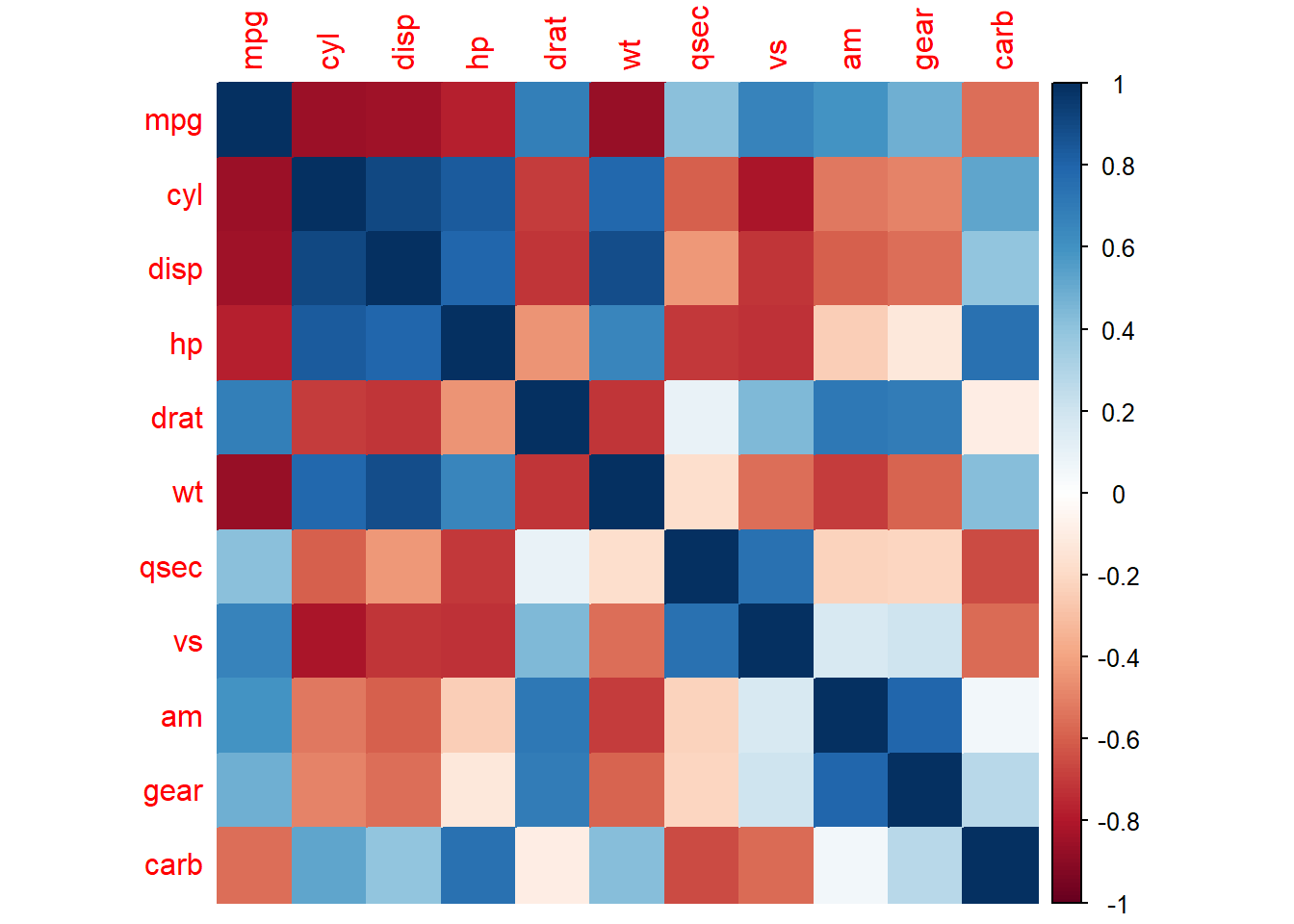
with clustering
corrplot::corrplot(M, order = "hclust")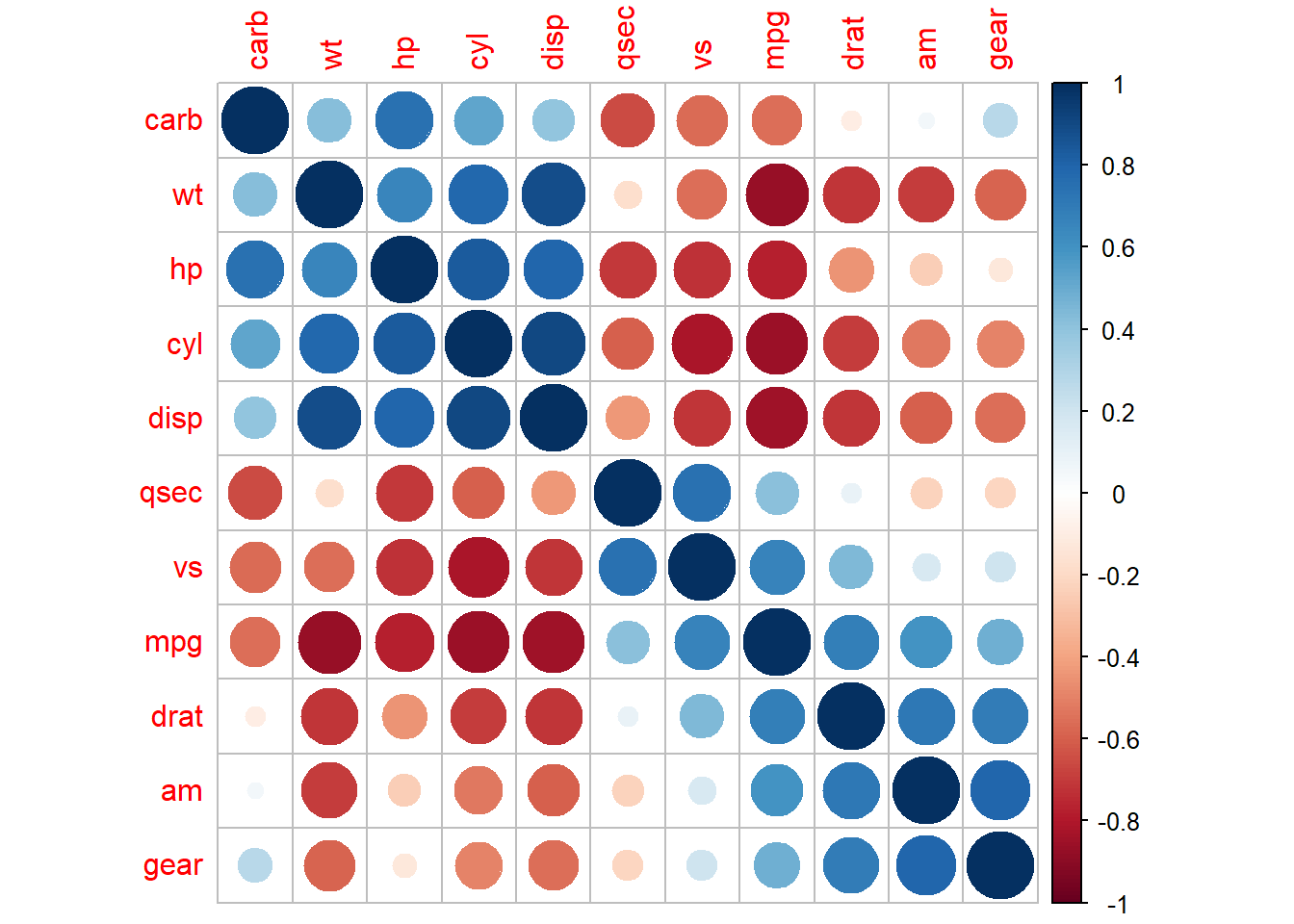
filter correlations at a cutoff value
caret::findCorrelation(M, cutoff = 0.2, verbose = TRUE, names = TRUE)
#> Compare row 2 and column 3 with corr 0.902
#> Means: 0.701 vs 0.546 so flagging column 2
#> Compare row 3 and column 1 with corr 0.848
#> Means: 0.658 vs 0.513 so flagging column 3
#> Compare row 1 and column 6 with corr 0.868
#> Means: 0.63 vs 0.483 so flagging column 1
#> Compare row 6 and column 4 with corr 0.659
#> Means: 0.543 vs 0.446 so flagging column 6
#> Compare row 4 and column 8 with corr 0.723
#> Means: 0.5 vs 0.418 so flagging column 4
#> Compare row 8 and column 5 with corr 0.44
#> Means: 0.426 vs 0.394 so flagging column 8
#> Compare row 5 and column 9 with corr 0.713
#> Means: 0.399 vs 0.365 so flagging column 5
#> Compare row 9 and column 10 with corr 0.794
#> Means: 0.36 vs 0.352 so flagging column 9
#> Compare row 10 and column 11 with corr 0.274
#> Means: 0.243 vs 0.339 so flagging column 11
#> Compare row 10 and column 7 with corr 0.213
#> Means: 0.213 vs 0.213 so flagging column 7
#> All correlations <= 0.2
#> [1] "cyl" "disp" "mpg" "wt" "hp" "vs" "drat" "am" "carb" "qsec"I dont really like the formatting here - too hard to match columns - let’s find a better way.
There is corrr::correlations() but not available on CRAN and there is also an open issue with the newest version of dplyr.
Using deprecrated reshape2 package because tidyr doesn’t handle matrixs. This is fine for now.
reshape2::melt(M) %>%
dplyr::filter(abs(value) > 0.2, Var1 != Var2) %>%
dplyr::arrange(desc(abs(value))) %>%
head()
#> Var1 Var2 value
#> 1 disp cyl 0.9020329
#> 2 cyl disp 0.9020329
#> 3 wt disp 0.8879799
#> 4 disp wt 0.8879799
#> 5 wt mpg -0.8676594
#> 6 mpg wt -0.8676594plot distribution of all variables
Just the numeric variables
SOURCE: (https://drsimonj.svbtle.com/quick-plot-of-all-variables)
mtcars %>%
purrr::keep(is.numeric) %>%
tidyr::gather() %>%
ggplot2::ggplot(ggplot2::aes(value)) +
ggplot2::facet_wrap(~key, scales = "free") +
ggplot2::geom_histogram()
#> `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.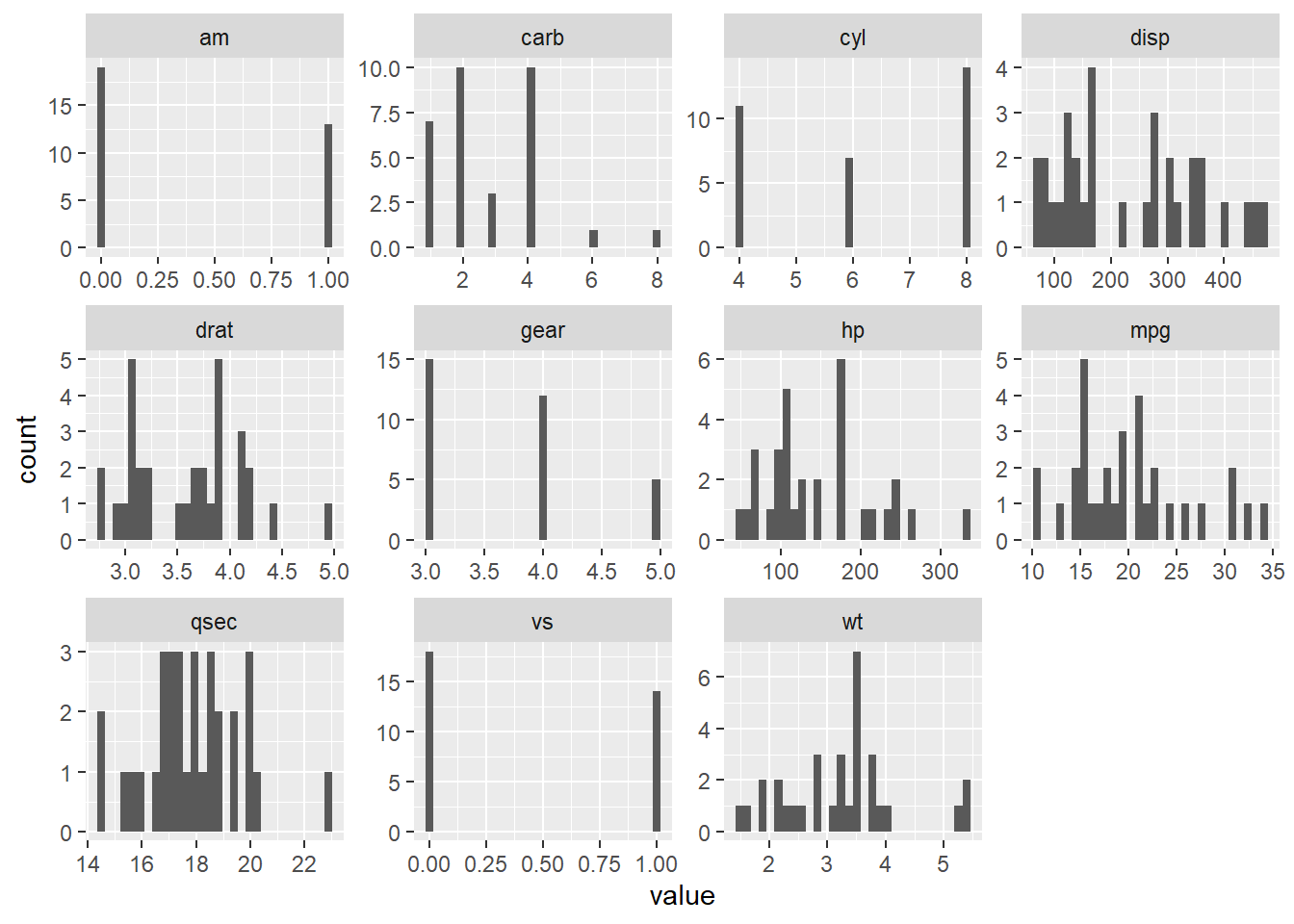
all variables
nycflights13::planes[,c(4,9)] %>%
tidyr::gather() %>%
ggplot2::ggplot(ggplot2::aes(value)) +
ggplot2::facet_wrap(~key, scales = "free") +
ggplot2::geom_bar(stat = "count") +
ggplot2::coord_flip ()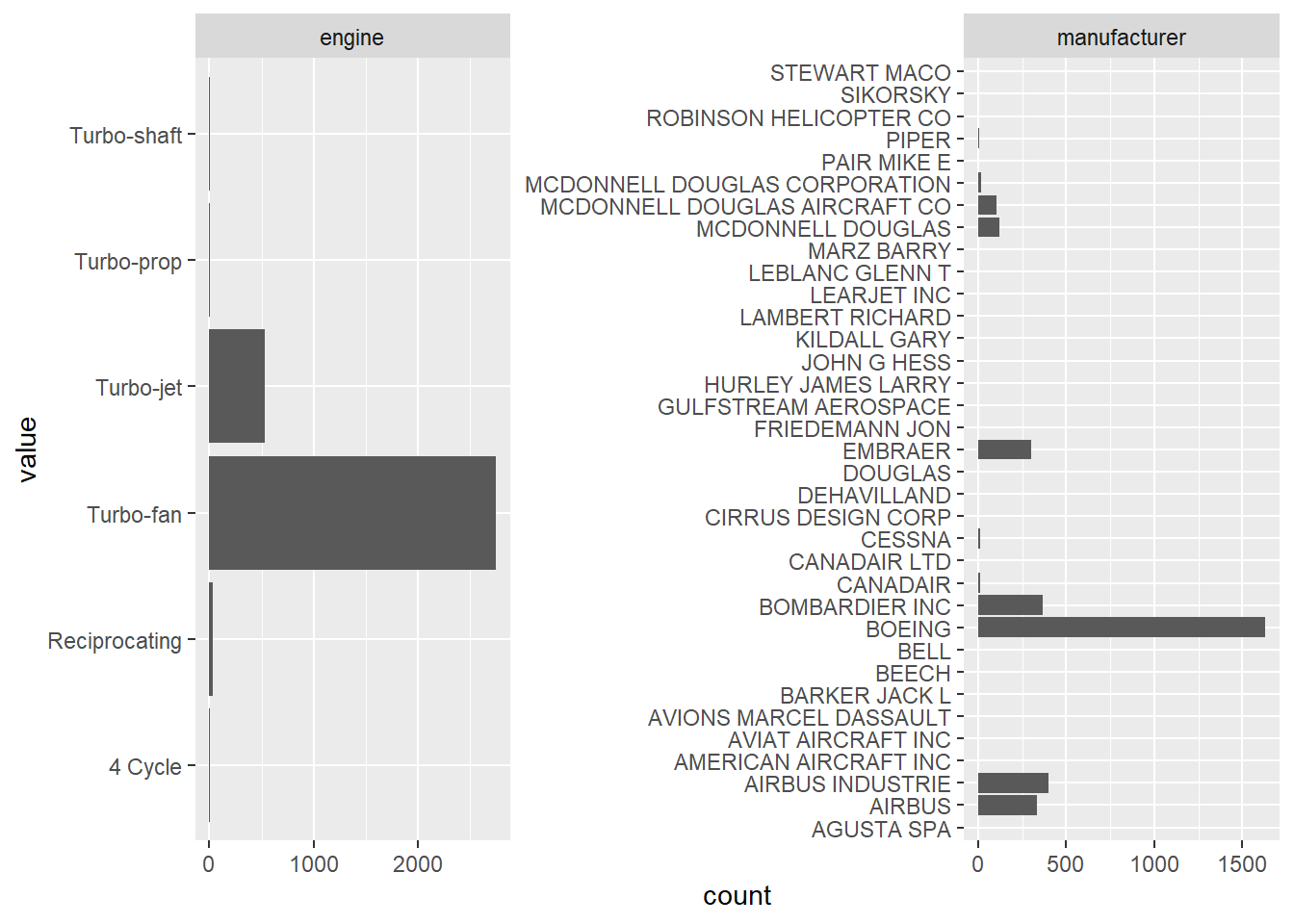
RMarkdown: insert date document was knitted
add this to the header:
SOURCE: (https://stackoverflow.com/questions/23449319/yaml-current-date-in-rmarkdown)
Plot percentage of attributes that are NA for each outcome
data(Soybean, package = "mlbench")
# the first column is the outcome variable so we remove it
na_rates <- rowMeans(is.na(Soybean[,-1]))
soybean <- Soybean %>%
tibble::add_column(na_rate = na_rates)
soybean %>%
dplyr::group_by(Class) %>%
dplyr::summarize(mean_na_rate = mean(na_rate)) %>%
ggplot2::ggplot(ggplot2::aes(x=Class, y = mean_na_rate, fill = Class)) +
ggplot2::geom_bar(stat = "identity") +
ggplot2::coord_flip() +
ggplot2::theme(legend.position = "none")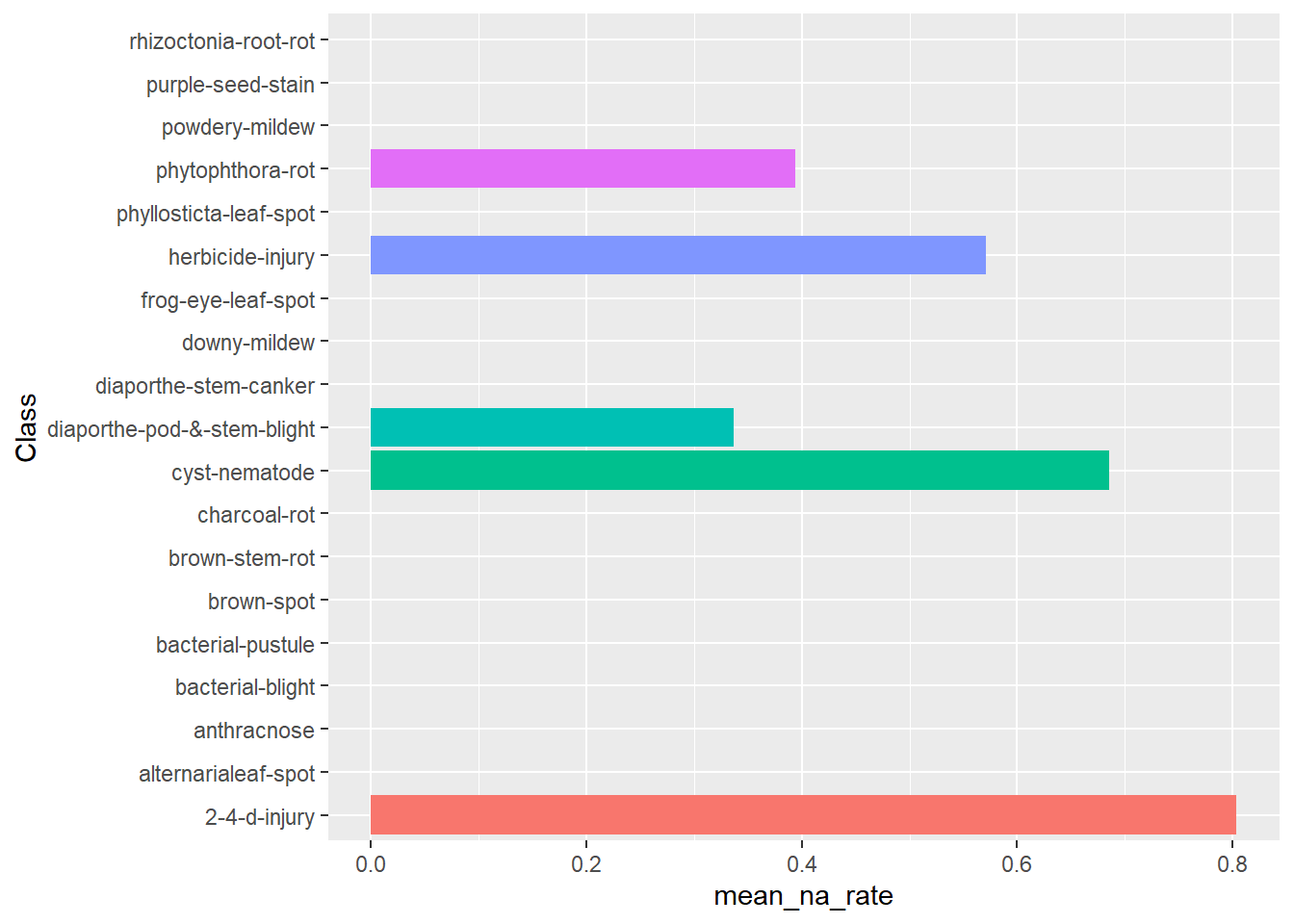
Plot the pecentage of rows that has at least 1 NA attribute, by outcome
data(Soybean, package = "mlbench")
na_rates <- rowMeans(is.na(Soybean[,-1]))
soybean <- Soybean %>%
tibble::add_column(na_rates = na_rates) %>%
dplyr::mutate(has_nas = na_rates != 0 )
soybean %>%
dplyr::group_by(Class) %>%
dplyr::summarize(mean_has_nas = mean(has_nas)) %>%
ggplot2::ggplot(ggplot2::aes(x=Class, y = mean_has_nas, fill = Class)) +
ggplot2::geom_bar(stat = "identity") +
ggplot2::coord_flip() +
ggplot2::theme(legend.position = "none")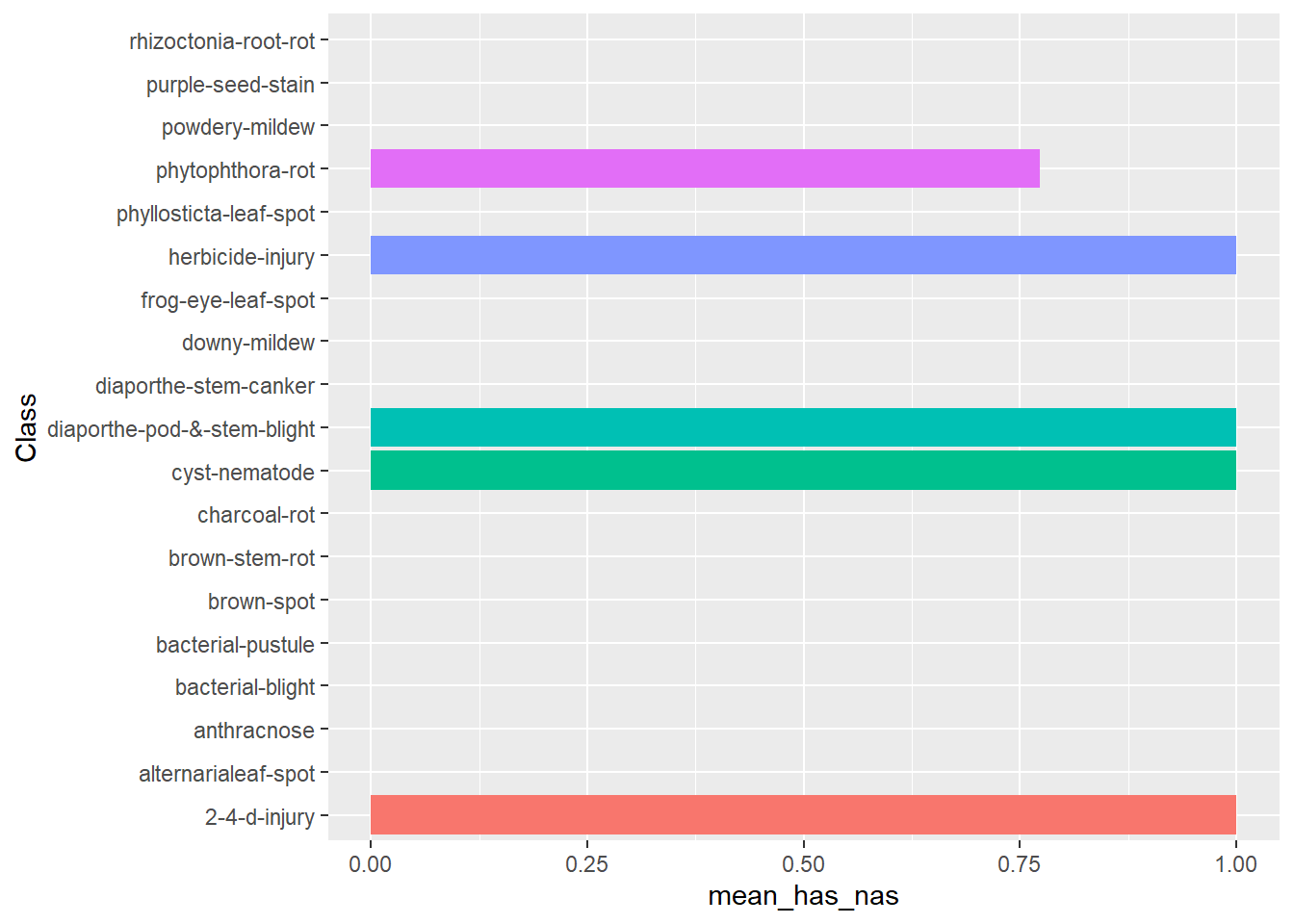
Plot the attributes (predictors) that are most likely to be missing
data(Soybean, package = "mlbench")
predictor_na_rate <- data.frame(colMeans(is.na(Soybean[,-1]))) %>%
tibble::rownames_to_column(var = "predictor") %>%
dplyr::rename( na_rate = 2)
predictor_na_rate %>% ggplot2::ggplot(ggplot2::aes(x = predictor, y = na_rate,
fill = predictor )) +
ggplot2::geom_bar(stat = "identity") +
ggplot2::coord_flip() +
ggplot2::theme(legend.position = "none")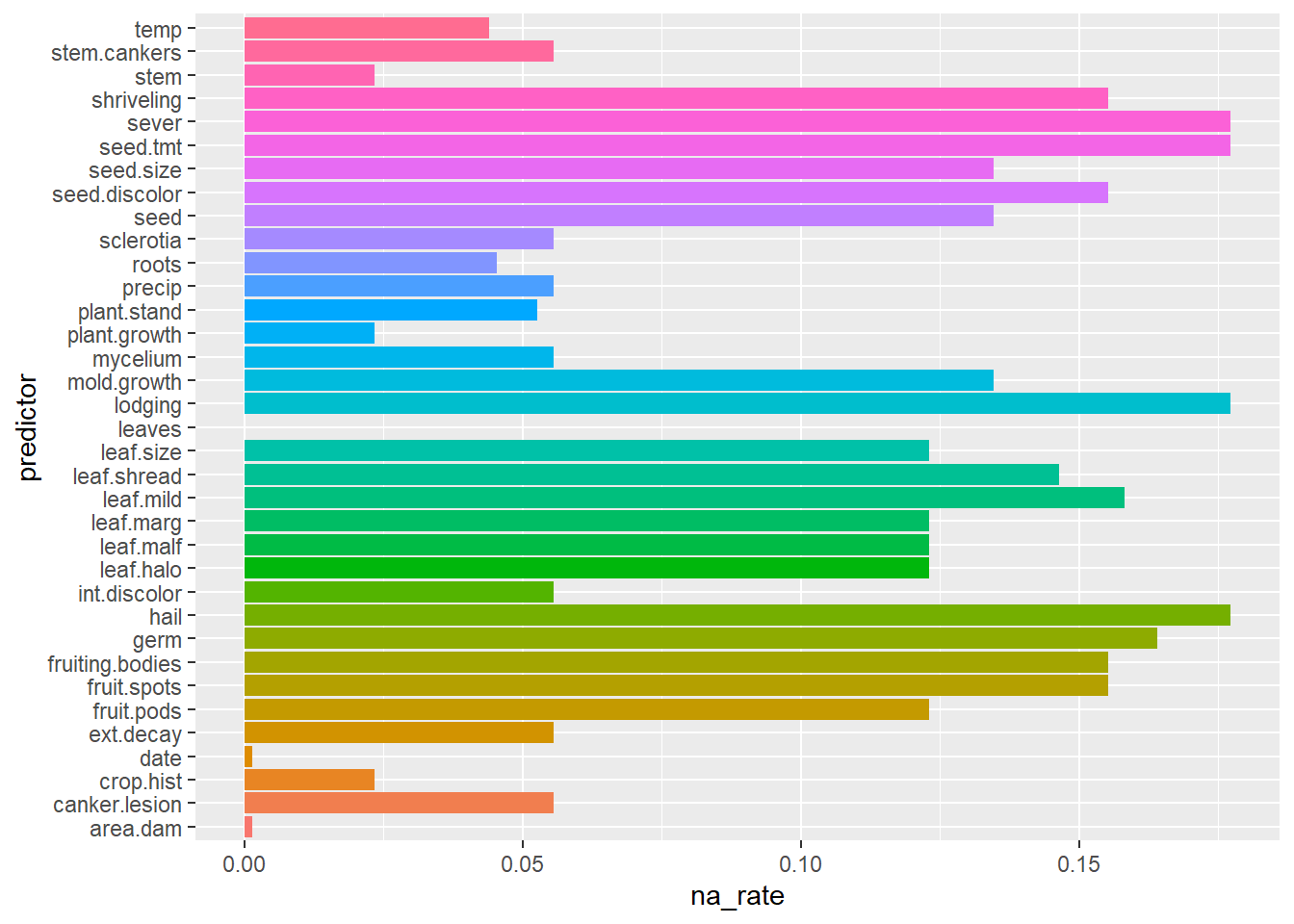
Plot the attributes (predictors) that are most likely to be missing, by outcome
data(Soybean, package = "mlbench")
soybean_l <- Soybean %>%
tidyr::gather(-Class, key = "predictor",value = "value")
#> Warning: attributes are not identical across measure variables;
#> they will be dropped
soybean_l %>% dplyr::group_by(Class, predictor) %>%
dplyr::summarize(na_rate = mean(is.na(value))) %>%
ggplot2::ggplot(ggplot2::aes(x=predictor, y = na_rate, fill = predictor)) +
ggplot2::geom_bar(stat = "identity") +
ggplot2::coord_flip() +
ggplot2::facet_wrap(~Class, scales = "free") +
ggplot2::theme(legend.position = "none")
#> `summarise()` has grouped output by 'Class'. You can override using the `.groups` argument.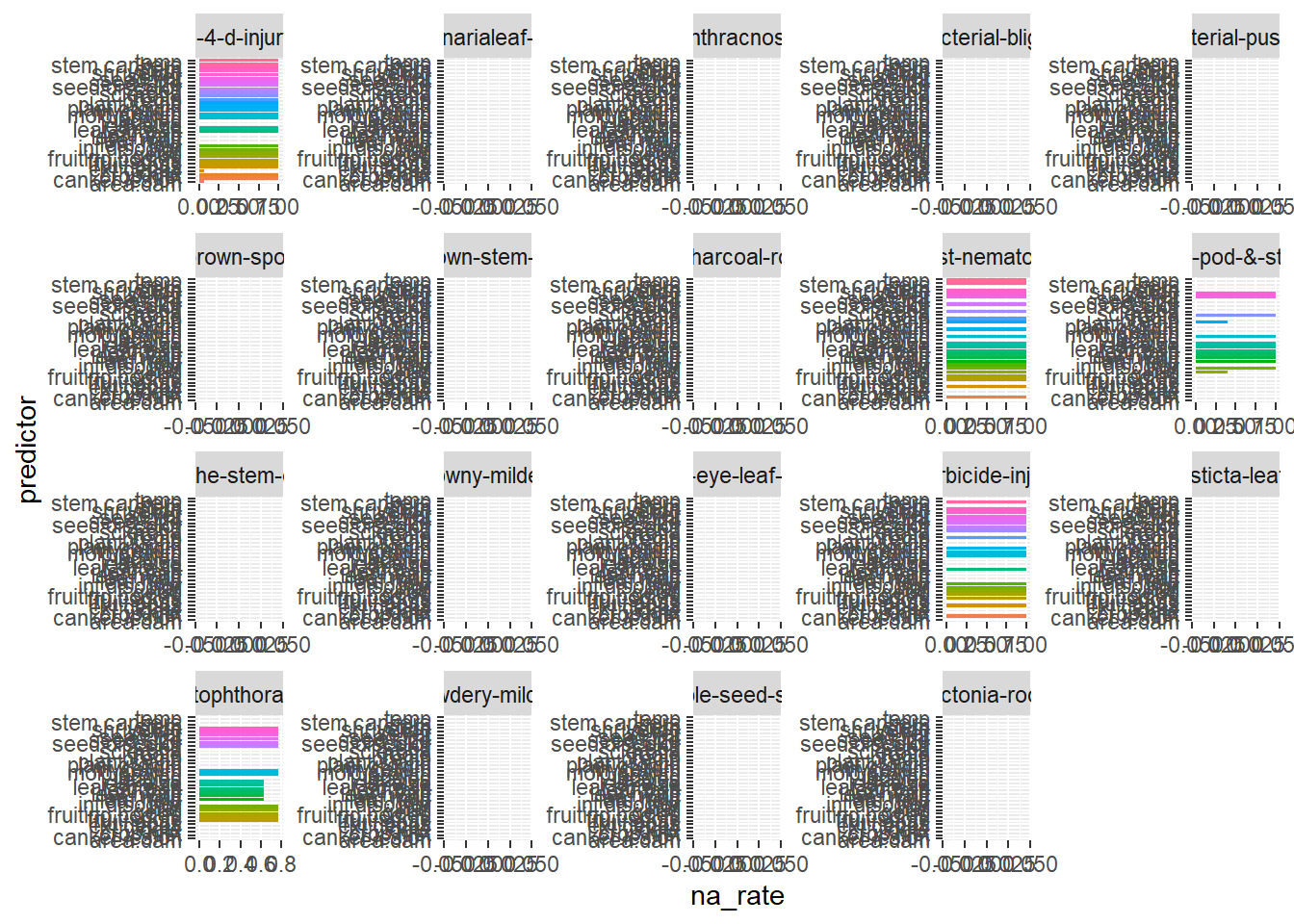
Create a matrix that shows whether or not a particular combination of values is in the data
SOURCE: https://stackoverflow.com/a/37897416
mtcars %>%
tibble::rownames_to_column("name") %>%
dplyr::distinct(cyl, gear, has_gear = !is.na(name)) %>%
tidyr::complete( cyl, gear) %>%
dplyr::mutate(has_gear = dplyr::if_else(is.na(has_gear),FALSE, TRUE)) %>%
ggplot2::ggplot(ggplot2::aes(x=cyl, y =gear )) +
ggplot2::geom_tile(ggplot2::aes(fill = has_gear),color = "white")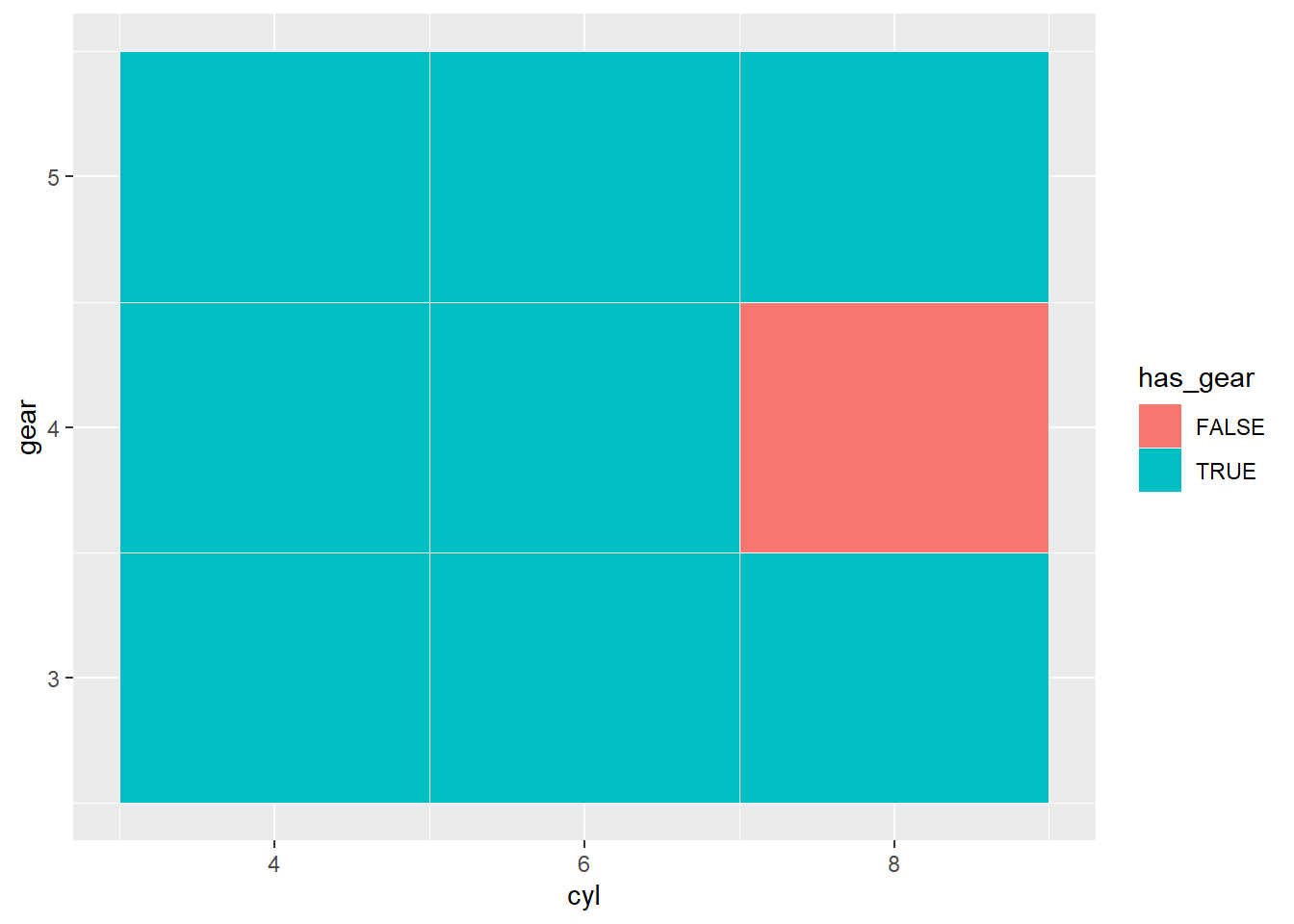
Convert unix style epoch time to human readable time
epoch_time <- 1505249866329
lubridate::as_datetime(epoch_time/1000)
#> [1] "2017-09-12 20:57:46 UTC"Clean Data: remove columns where no rows contain a value
df <-data.frame(col_1 = c(NA, 1, 2, 3),
col_2 = c(NA, NA, NA, NA),
col_3 = c(1, 2, 3, 4))
df
#> col_1 col_2 col_3
#> 1 NA NA 1
#> 2 1 NA 2
#> 3 2 NA 3
#> 4 3 NA 4
case_missing_rate <- data.frame(colMeans(is.na(df))) %>%
tibble::rownames_to_column(var = "column") %>%
dplyr::rename(missing_rate = 2)
all_missing_cols <- case_missing_rate %>%
dplyr::filter(missing_rate == 1.0) %>%
dplyr::pull(column)
df %>%
dplyr::select(-all_missing_cols)
#> Note: Using an external vector in selections is ambiguous.
#> i Use `all_of(all_missing_cols)` instead of `all_missing_cols` to silence this message.
#> i See <https://tidyselect.r-lib.org/reference/faq-external-vector.html>.
#> This message is displayed once per session.
#> col_1 col_3
#> 1 NA 1
#> 2 1 2
#> 3 2 3
#> 4 3 4Print one plot for each data frame in a list column
There is probably a better way to meet the need.
My goals was to only show the values that are present in a given group.
ggplot2::facet_wrap() shows all values present in any group in the plots for
all groups.
Step 1: make the list
n_mtcars <-
mtcars %>%
dplyr::group_by(cyl) %>%
tidyr::nest()
n_mtcars
#> # A tibble: 3 x 2
#> # Groups: cyl [3]
#> cyl data
#> <dbl> <list>
#> 1 6 <tibble [7 x 10]>
#> 2 4 <tibble [11 x 10]>
#> 3 8 <tibble [14 x 10]>Step 2: give the list members names
names(n_mtcars$data) <- stringr::str_c(n_mtcars$cyl,"_cyl")
str(n_mtcars$data)
#> List of 3
#> $ 6_cyl: tibble [7 x 10] (S3: tbl_df/tbl/data.frame)
#> ..$ mpg : num [1:7] 21 21 21.4 18.1 19.2 17.8 19.7
#> ..$ disp: num [1:7] 160 160 258 225 168 ...
#> ..$ hp : num [1:7] 110 110 110 105 123 123 175
#> ..$ drat: num [1:7] 3.9 3.9 3.08 2.76 3.92 3.92 3.62
#> ..$ wt : num [1:7] 2.62 2.88 3.21 3.46 3.44 ...
#> ..$ qsec: num [1:7] 16.5 17 19.4 20.2 18.3 ...
#> ..$ vs : num [1:7] 0 0 1 1 1 1 0
#> ..$ am : num [1:7] 1 1 0 0 0 0 1
#> ..$ gear: num [1:7] 4 4 3 3 4 4 5
#> ..$ carb: num [1:7] 4 4 1 1 4 4 6
#> $ 4_cyl: tibble [11 x 10] (S3: tbl_df/tbl/data.frame)
#> ..$ mpg : num [1:11] 22.8 24.4 22.8 32.4 30.4 33.9 21.5 27.3 26 30.4 ...
#> ..$ disp: num [1:11] 108 146.7 140.8 78.7 75.7 ...
#> ..$ hp : num [1:11] 93 62 95 66 52 65 97 66 91 113 ...
#> ..$ drat: num [1:11] 3.85 3.69 3.92 4.08 4.93 4.22 3.7 4.08 4.43 3.77 ...
#> ..$ wt : num [1:11] 2.32 3.19 3.15 2.2 1.61 ...
#> ..$ qsec: num [1:11] 18.6 20 22.9 19.5 18.5 ...
#> ..$ vs : num [1:11] 1 1 1 1 1 1 1 1 0 1 ...
#> ..$ am : num [1:11] 1 0 0 1 1 1 0 1 1 1 ...
#> ..$ gear: num [1:11] 4 4 4 4 4 4 3 4 5 5 ...
#> ..$ carb: num [1:11] 1 2 2 1 2 1 1 1 2 2 ...
#> $ 8_cyl: tibble [14 x 10] (S3: tbl_df/tbl/data.frame)
#> ..$ mpg : num [1:14] 18.7 14.3 16.4 17.3 15.2 10.4 10.4 14.7 15.5 15.2 ...
#> ..$ disp: num [1:14] 360 360 276 276 276 ...
#> ..$ hp : num [1:14] 175 245 180 180 180 205 215 230 150 150 ...
#> ..$ drat: num [1:14] 3.15 3.21 3.07 3.07 3.07 2.93 3 3.23 2.76 3.15 ...
#> ..$ wt : num [1:14] 3.44 3.57 4.07 3.73 3.78 ...
#> ..$ qsec: num [1:14] 17 15.8 17.4 17.6 18 ...
#> ..$ vs : num [1:14] 0 0 0 0 0 0 0 0 0 0 ...
#> ..$ am : num [1:14] 0 0 0 0 0 0 0 0 0 0 ...
#> ..$ gear: num [1:14] 3 3 3 3 3 3 3 3 3 3 ...
#> ..$ carb: num [1:14] 2 4 3 3 3 4 4 4 2 2 ...Step 3: Create plotting function
plot_issues_by_epic <- function(df, item_name) {
df %>%
ggplot2::ggplot(ggplot2::aes(gear)) +
ggplot2::geom_bar() +
ggplot2::coord_flip() +
ggplot2::labs(title= item_name)
}Step 4: Use your plotting function to create the plots
purrr::map2(n_mtcars$data, names(n_mtcars$data),
plot_issues_by_epic)
#> $`6_cyl`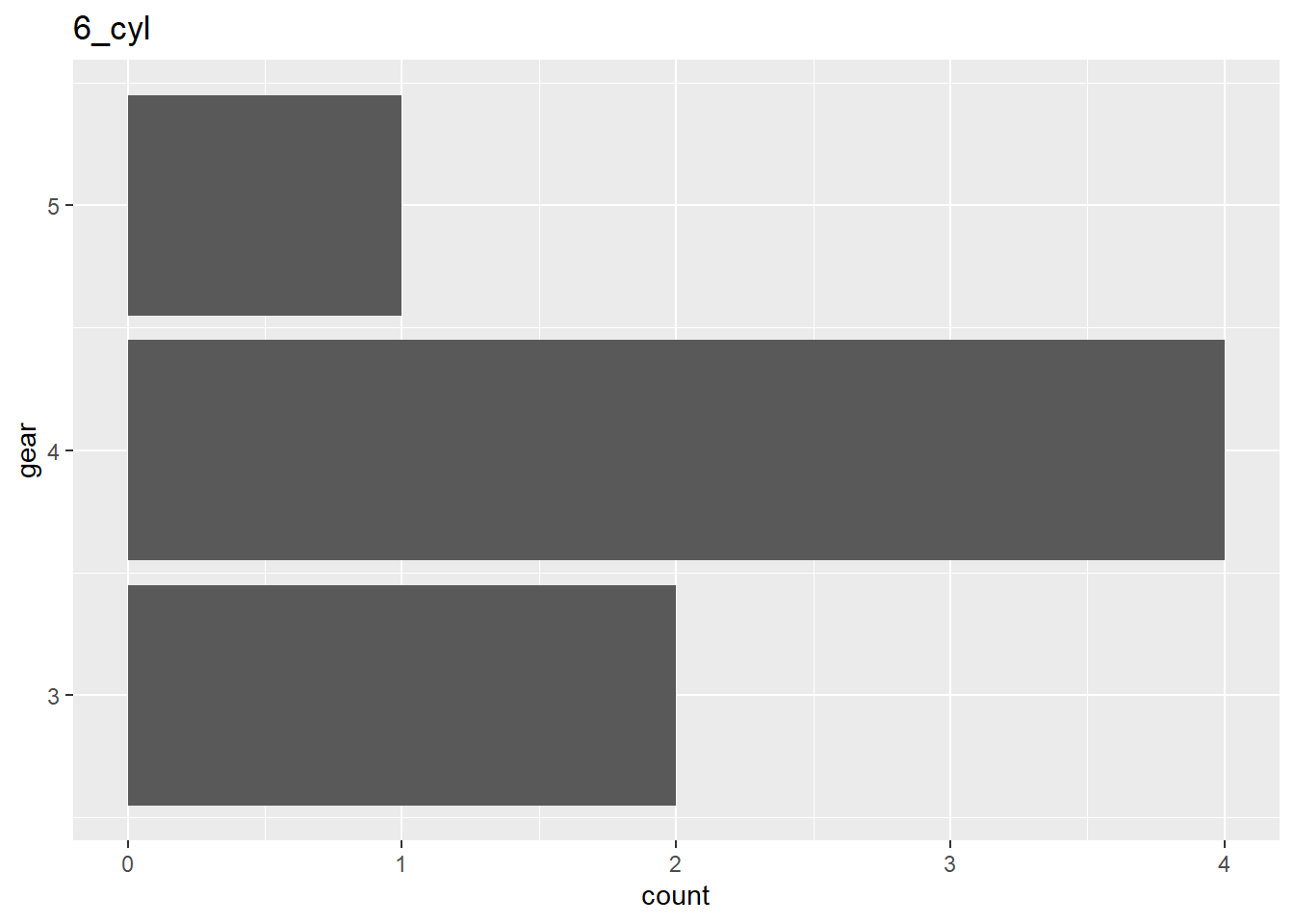
#>
#> $`4_cyl`
#>
#> $`8_cyl`
One plot for each attribute in a data frame - different scale for each attribute
datasets::airquality %>%
dplyr::filter(Month == 5) %>%
dplyr::select(-Month) %>%
tidyr::pivot_longer(-Day, names_to = "attribute", values_to = "value") %>%
ggplot2::ggplot(ggplot2::aes(Day, value)) +
ggplot2::geom_line() +
ggplot2::facet_grid(attribute ~ ., scales = "free")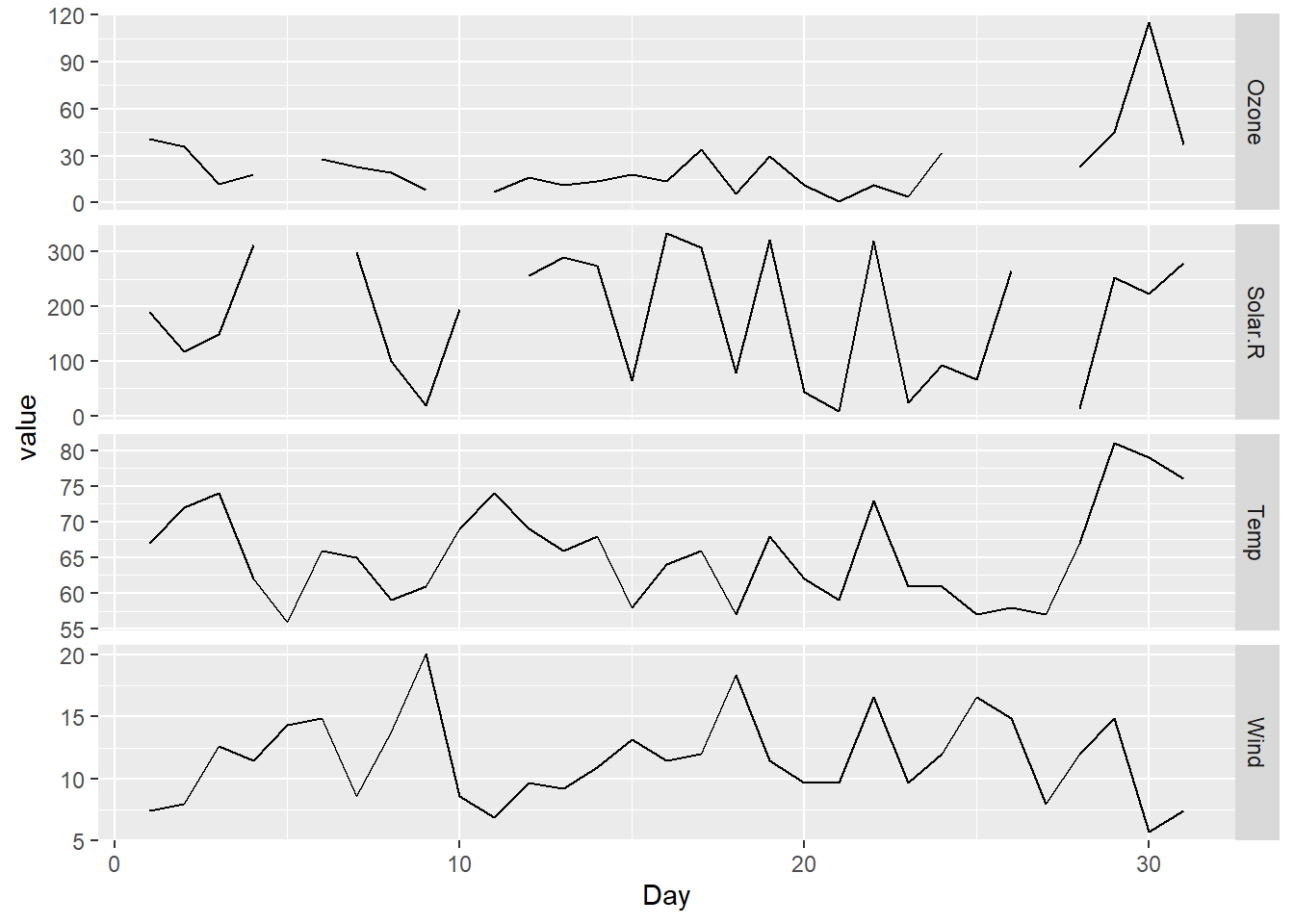
Get detailed information for each column in a database table
odbc::odbcConnectionColumns(con, "table_name")Get just the columns and datatypes for each column in a database table
odbc::odbcListColumns(con, table = "table_name")Get just the column names
DBI::dbListFields(con, "table_name")Table of contents for rmarkdown document
Add this to the document header.
output:
html_document:
toc: true
toc_depth: 6
toc_float: trueGet unique values from all non-numeric columns
dplyr::starwars %>%
# this works for list, but removing them to make the example output
# more concise
dplyr::select(!where(is.numeric) & !where(is.list)) %>%
purrr::map(unique)
#> $name
#> [1] "Luke Skywalker" "C-3PO" "R2-D2"
#> [4] "Darth Vader" "Leia Organa" "Owen Lars"
#> [7] "Beru Whitesun lars" "R5-D4" "Biggs Darklighter"
#> [10] "Obi-Wan Kenobi" "Anakin Skywalker" "Wilhuff Tarkin"
#> [13] "Chewbacca" "Han Solo" "Greedo"
#> [16] "Jabba Desilijic Tiure" "Wedge Antilles" "Jek Tono Porkins"
#> [19] "Yoda" "Palpatine" "Boba Fett"
#> [22] "IG-88" "Bossk" "Lando Calrissian"
#> [25] "Lobot" "Ackbar" "Mon Mothma"
#> [28] "Arvel Crynyd" "Wicket Systri Warrick" "Nien Nunb"
#> [31] "Qui-Gon Jinn" "Nute Gunray" "Finis Valorum"
#> [34] "Jar Jar Binks" "Roos Tarpals" "Rugor Nass"
#> [37] "Ric Olié" "Watto" "Sebulba"
#> [40] "Quarsh Panaka" "Shmi Skywalker" "Darth Maul"
#> [43] "Bib Fortuna" "Ayla Secura" "Dud Bolt"
#> [46] "Gasgano" "Ben Quadinaros" "Mace Windu"
#> [49] "Ki-Adi-Mundi" "Kit Fisto" "Eeth Koth"
#> [52] "Adi Gallia" "Saesee Tiin" "Yarael Poof"
#> [55] "Plo Koon" "Mas Amedda" "Gregar Typho"
#> [58] "Cordé" "Cliegg Lars" "Poggle the Lesser"
#> [61] "Luminara Unduli" "Barriss Offee" "Dormé"
#> [64] "Dooku" "Bail Prestor Organa" "Jango Fett"
#> [67] "Zam Wesell" "Dexter Jettster" "Lama Su"
#> [70] "Taun We" "Jocasta Nu" "Ratts Tyerell"
#> [73] "R4-P17" "Wat Tambor" "San Hill"
#> [76] "Shaak Ti" "Grievous" "Tarfful"
#> [79] "Raymus Antilles" "Sly Moore" "Tion Medon"
#> [82] "Finn" "Rey" "Poe Dameron"
#> [85] "BB8" "Captain Phasma" "Padmé Amidala"
#>
#> $hair_color
#> [1] "blond" NA "none" "brown"
#> [5] "brown, grey" "black" "auburn, white" "auburn, grey"
#> [9] "white" "grey" "auburn" "blonde"
#> [13] "unknown"
#>
#> $skin_color
#> [1] "fair" "gold" "white, blue"
#> [4] "white" "light" "white, red"
#> [7] "unknown" "green" "green-tan, brown"
#> [10] "pale" "metal" "dark"
#> [13] "brown mottle" "brown" "grey"
#> [16] "mottled green" "orange" "blue, grey"
#> [19] "grey, red" "red" "blue"
#> [22] "grey, green, yellow" "yellow" "tan"
#> [25] "fair, green, yellow" "grey, blue" "silver, red"
#> [28] "green, grey" "red, blue, white" "brown, white"
#> [31] "none"
#>
#> $eye_color
#> [1] "blue" "yellow" "red" "brown"
#> [5] "blue-gray" "black" "orange" "hazel"
#> [9] "pink" "unknown" "red, blue" "gold"
#> [13] "green, yellow" "white" "dark"
#>
#> $sex
#> [1] "male" "none" "female" "hermaphroditic"
#> [5] NA
#>
#> $gender
#> [1] "masculine" "feminine" NA
#>
#> $homeworld
#> [1] "Tatooine" "Naboo" "Alderaan" "Stewjon"
#> [5] "Eriadu" "Kashyyyk" "Corellia" "Rodia"
#> [9] "Nal Hutta" "Bestine IV" NA "Kamino"
#> [13] "Trandosha" "Socorro" "Bespin" "Mon Cala"
#> [17] "Chandrila" "Endor" "Sullust" "Cato Neimoidia"
#> [21] "Coruscant" "Toydaria" "Malastare" "Dathomir"
#> [25] "Ryloth" "Vulpter" "Troiken" "Tund"
#> [29] "Haruun Kal" "Cerea" "Glee Anselm" "Iridonia"
#> [33] "Iktotch" "Quermia" "Dorin" "Champala"
#> [37] "Geonosis" "Mirial" "Serenno" "Concord Dawn"
#> [41] "Zolan" "Ojom" "Aleen Minor" "Skako"
#> [45] "Muunilinst" "Shili" "Kalee" "Umbara"
#> [49] "Utapau"
#>
#> $species
#> [1] "Human" "Droid" "Wookiee" "Rodian"
#> [5] "Hutt" "Yoda's species" "Trandoshan" "Mon Calamari"
#> [9] "Ewok" "Sullustan" "Neimodian" "Gungan"
#> [13] NA "Toydarian" "Dug" "Zabrak"
#> [17] "Twi'lek" "Vulptereen" "Xexto" "Toong"
#> [21] "Cerean" "Nautolan" "Tholothian" "Iktotchi"
#> [25] "Quermian" "Kel Dor" "Chagrian" "Geonosian"
#> [29] "Mirialan" "Clawdite" "Besalisk" "Kaminoan"
#> [33] "Aleena" "Skakoan" "Muun" "Togruta"
#> [37] "Kaleesh" "Pau'an"Get count and proportion of all non-numeric columns
Counts…
dplyr::starwars %>%
# this works for list, but removing them to make the example output
# more concise
dplyr::select(!where(is.numeric) & !where(is.list)) %>%
purrr::map(~ table(.))
#> $name
#> .
#> Ackbar Adi Gallia Anakin Skywalker
#> 1 1 1
#> Arvel Crynyd Ayla Secura Bail Prestor Organa
#> 1 1 1
#> Barriss Offee BB8 Ben Quadinaros
#> 1 1 1
#> Beru Whitesun lars Bib Fortuna Biggs Darklighter
#> 1 1 1
#> Boba Fett Bossk C-3PO
#> 1 1 1
#> Captain Phasma Chewbacca Cliegg Lars
#> 1 1 1
#> Cordé Darth Maul Darth Vader
#> 1 1 1
#> Dexter Jettster Dooku Dormé
#> 1 1 1
#> Dud Bolt Eeth Koth Finis Valorum
#> 1 1 1
#> Finn Gasgano Greedo
#> 1 1 1
#> Gregar Typho Grievous Han Solo
#> 1 1 1
#> IG-88 Jabba Desilijic Tiure Jango Fett
#> 1 1 1
#> Jar Jar Binks Jek Tono Porkins Jocasta Nu
#> 1 1 1
#> Ki-Adi-Mundi Kit Fisto Lama Su
#> 1 1 1
#> Lando Calrissian Leia Organa Lobot
#> 1 1 1
#> Luke Skywalker Luminara Unduli Mace Windu
#> 1 1 1
#> Mas Amedda Mon Mothma Nien Nunb
#> 1 1 1
#> Nute Gunray Obi-Wan Kenobi Owen Lars
#> 1 1 1
#> Padmé Amidala Palpatine Plo Koon
#> 1 1 1
#> Poe Dameron Poggle the Lesser Quarsh Panaka
#> 1 1 1
#> Qui-Gon Jinn R2-D2 R4-P17
#> 1 1 1
#> R5-D4 Ratts Tyerell Raymus Antilles
#> 1 1 1
#> Rey Ric Olié Roos Tarpals
#> 1 1 1
#> Rugor Nass Saesee Tiin San Hill
#> 1 1 1
#> Sebulba Shaak Ti Shmi Skywalker
#> 1 1 1
#> Sly Moore Tarfful Taun We
#> 1 1 1
#> Tion Medon Wat Tambor Watto
#> 1 1 1
#> Wedge Antilles Wicket Systri Warrick Wilhuff Tarkin
#> 1 1 1
#> Yarael Poof Yoda Zam Wesell
#> 1 1 1
#>
#> $hair_color
#> .
#> auburn auburn, grey auburn, white black blond
#> 1 1 1 13 3
#> blonde brown brown, grey grey none
#> 1 18 1 1 37
#> unknown white
#> 1 4
#>
#> $skin_color
#> .
#> blue blue, grey brown brown mottle
#> 2 2 4 1
#> brown, white dark fair fair, green, yellow
#> 1 6 17 1
#> gold green green-tan, brown green, grey
#> 1 6 1 1
#> grey grey, blue grey, green, yellow grey, red
#> 6 1 1 1
#> light metal mottled green none
#> 11 1 1 1
#> orange pale red red, blue, white
#> 2 5 1 1
#> silver, red tan unknown white
#> 1 2 2 2
#> white, blue white, red yellow
#> 2 1 2
#>
#> $eye_color
#> .
#> black blue blue-gray brown dark
#> 10 19 1 21 1
#> gold green, yellow hazel orange pink
#> 1 1 3 8 1
#> red red, blue unknown white yellow
#> 5 1 3 1 11
#>
#> $sex
#> .
#> female hermaphroditic male none
#> 16 1 60 6
#>
#> $gender
#> .
#> feminine masculine
#> 17 66
#>
#> $homeworld
#> .
#> Alderaan Aleen Minor Bespin Bestine IV Cato Neimoidia
#> 3 1 1 1 1
#> Cerea Champala Chandrila Concord Dawn Corellia
#> 1 1 1 1 2
#> Coruscant Dathomir Dorin Endor Eriadu
#> 3 1 1 1 1
#> Geonosis Glee Anselm Haruun Kal Iktotch Iridonia
#> 1 1 1 1 1
#> Kalee Kamino Kashyyyk Malastare Mirial
#> 1 3 2 1 2
#> Mon Cala Muunilinst Naboo Nal Hutta Ojom
#> 1 1 11 1 1
#> Quermia Rodia Ryloth Serenno Shili
#> 1 1 2 1 1
#> Skako Socorro Stewjon Sullust Tatooine
#> 1 1 1 1 10
#> Toydaria Trandosha Troiken Tund Umbara
#> 1 1 1 1 1
#> Utapau Vulpter Zolan
#> 1 1 1
#>
#> $species
#> .
#> Aleena Besalisk Cerean Chagrian Clawdite
#> 1 1 1 1 1
#> Droid Dug Ewok Geonosian Gungan
#> 6 1 1 1 3
#> Human Hutt Iktotchi Kaleesh Kaminoan
#> 35 1 1 1 2
#> Kel Dor Mirialan Mon Calamari Muun Nautolan
#> 1 2 1 1 1
#> Neimodian Pau'an Quermian Rodian Skakoan
#> 1 1 1 1 1
#> Sullustan Tholothian Togruta Toong Toydarian
#> 1 1 1 1 1
#> Trandoshan Twi'lek Vulptereen Wookiee Xexto
#> 1 2 1 2 1
#> Yoda's species Zabrak
#> 1 2Proportions…
dplyr::starwars %>%
# this works for list, but removing them to make the example output
# more concise
dplyr::select(!where(is.numeric) & !where(is.list)) %>%
purrr::map(~ table(.) %>% prop.table())
#> $name
#> .
#> Ackbar Adi Gallia Anakin Skywalker
#> 0.01149425 0.01149425 0.01149425
#> Arvel Crynyd Ayla Secura Bail Prestor Organa
#> 0.01149425 0.01149425 0.01149425
#> Barriss Offee BB8 Ben Quadinaros
#> 0.01149425 0.01149425 0.01149425
#> Beru Whitesun lars Bib Fortuna Biggs Darklighter
#> 0.01149425 0.01149425 0.01149425
#> Boba Fett Bossk C-3PO
#> 0.01149425 0.01149425 0.01149425
#> Captain Phasma Chewbacca Cliegg Lars
#> 0.01149425 0.01149425 0.01149425
#> Cordé Darth Maul Darth Vader
#> 0.01149425 0.01149425 0.01149425
#> Dexter Jettster Dooku Dormé
#> 0.01149425 0.01149425 0.01149425
#> Dud Bolt Eeth Koth Finis Valorum
#> 0.01149425 0.01149425 0.01149425
#> Finn Gasgano Greedo
#> 0.01149425 0.01149425 0.01149425
#> Gregar Typho Grievous Han Solo
#> 0.01149425 0.01149425 0.01149425
#> IG-88 Jabba Desilijic Tiure Jango Fett
#> 0.01149425 0.01149425 0.01149425
#> Jar Jar Binks Jek Tono Porkins Jocasta Nu
#> 0.01149425 0.01149425 0.01149425
#> Ki-Adi-Mundi Kit Fisto Lama Su
#> 0.01149425 0.01149425 0.01149425
#> Lando Calrissian Leia Organa Lobot
#> 0.01149425 0.01149425 0.01149425
#> Luke Skywalker Luminara Unduli Mace Windu
#> 0.01149425 0.01149425 0.01149425
#> Mas Amedda Mon Mothma Nien Nunb
#> 0.01149425 0.01149425 0.01149425
#> Nute Gunray Obi-Wan Kenobi Owen Lars
#> 0.01149425 0.01149425 0.01149425
#> Padmé Amidala Palpatine Plo Koon
#> 0.01149425 0.01149425 0.01149425
#> Poe Dameron Poggle the Lesser Quarsh Panaka
#> 0.01149425 0.01149425 0.01149425
#> Qui-Gon Jinn R2-D2 R4-P17
#> 0.01149425 0.01149425 0.01149425
#> R5-D4 Ratts Tyerell Raymus Antilles
#> 0.01149425 0.01149425 0.01149425
#> Rey Ric Olié Roos Tarpals
#> 0.01149425 0.01149425 0.01149425
#> Rugor Nass Saesee Tiin San Hill
#> 0.01149425 0.01149425 0.01149425
#> Sebulba Shaak Ti Shmi Skywalker
#> 0.01149425 0.01149425 0.01149425
#> Sly Moore Tarfful Taun We
#> 0.01149425 0.01149425 0.01149425
#> Tion Medon Wat Tambor Watto
#> 0.01149425 0.01149425 0.01149425
#> Wedge Antilles Wicket Systri Warrick Wilhuff Tarkin
#> 0.01149425 0.01149425 0.01149425
#> Yarael Poof Yoda Zam Wesell
#> 0.01149425 0.01149425 0.01149425
#>
#> $hair_color
#> .
#> auburn auburn, grey auburn, white black blond
#> 0.01219512 0.01219512 0.01219512 0.15853659 0.03658537
#> blonde brown brown, grey grey none
#> 0.01219512 0.21951220 0.01219512 0.01219512 0.45121951
#> unknown white
#> 0.01219512 0.04878049
#>
#> $skin_color
#> .
#> blue blue, grey brown brown mottle
#> 0.02298851 0.02298851 0.04597701 0.01149425
#> brown, white dark fair fair, green, yellow
#> 0.01149425 0.06896552 0.19540230 0.01149425
#> gold green green-tan, brown green, grey
#> 0.01149425 0.06896552 0.01149425 0.01149425
#> grey grey, blue grey, green, yellow grey, red
#> 0.06896552 0.01149425 0.01149425 0.01149425
#> light metal mottled green none
#> 0.12643678 0.01149425 0.01149425 0.01149425
#> orange pale red red, blue, white
#> 0.02298851 0.05747126 0.01149425 0.01149425
#> silver, red tan unknown white
#> 0.01149425 0.02298851 0.02298851 0.02298851
#> white, blue white, red yellow
#> 0.02298851 0.01149425 0.02298851
#>
#> $eye_color
#> .
#> black blue blue-gray brown dark
#> 0.11494253 0.21839080 0.01149425 0.24137931 0.01149425
#> gold green, yellow hazel orange pink
#> 0.01149425 0.01149425 0.03448276 0.09195402 0.01149425
#> red red, blue unknown white yellow
#> 0.05747126 0.01149425 0.03448276 0.01149425 0.12643678
#>
#> $sex
#> .
#> female hermaphroditic male none
#> 0.19277108 0.01204819 0.72289157 0.07228916
#>
#> $gender
#> .
#> feminine masculine
#> 0.2048193 0.7951807
#>
#> $homeworld
#> .
#> Alderaan Aleen Minor Bespin Bestine IV Cato Neimoidia
#> 0.03896104 0.01298701 0.01298701 0.01298701 0.01298701
#> Cerea Champala Chandrila Concord Dawn Corellia
#> 0.01298701 0.01298701 0.01298701 0.01298701 0.02597403
#> Coruscant Dathomir Dorin Endor Eriadu
#> 0.03896104 0.01298701 0.01298701 0.01298701 0.01298701
#> Geonosis Glee Anselm Haruun Kal Iktotch Iridonia
#> 0.01298701 0.01298701 0.01298701 0.01298701 0.01298701
#> Kalee Kamino Kashyyyk Malastare Mirial
#> 0.01298701 0.03896104 0.02597403 0.01298701 0.02597403
#> Mon Cala Muunilinst Naboo Nal Hutta Ojom
#> 0.01298701 0.01298701 0.14285714 0.01298701 0.01298701
#> Quermia Rodia Ryloth Serenno Shili
#> 0.01298701 0.01298701 0.02597403 0.01298701 0.01298701
#> Skako Socorro Stewjon Sullust Tatooine
#> 0.01298701 0.01298701 0.01298701 0.01298701 0.12987013
#> Toydaria Trandosha Troiken Tund Umbara
#> 0.01298701 0.01298701 0.01298701 0.01298701 0.01298701
#> Utapau Vulpter Zolan
#> 0.01298701 0.01298701 0.01298701
#>
#> $species
#> .
#> Aleena Besalisk Cerean Chagrian Clawdite
#> 0.01204819 0.01204819 0.01204819 0.01204819 0.01204819
#> Droid Dug Ewok Geonosian Gungan
#> 0.07228916 0.01204819 0.01204819 0.01204819 0.03614458
#> Human Hutt Iktotchi Kaleesh Kaminoan
#> 0.42168675 0.01204819 0.01204819 0.01204819 0.02409639
#> Kel Dor Mirialan Mon Calamari Muun Nautolan
#> 0.01204819 0.02409639 0.01204819 0.01204819 0.01204819
#> Neimodian Pau'an Quermian Rodian Skakoan
#> 0.01204819 0.01204819 0.01204819 0.01204819 0.01204819
#> Sullustan Tholothian Togruta Toong Toydarian
#> 0.01204819 0.01204819 0.01204819 0.01204819 0.01204819
#> Trandoshan Twi'lek Vulptereen Wookiee Xexto
#> 0.01204819 0.02409639 0.01204819 0.02409639 0.01204819
#> Yoda's species Zabrak
#> 0.01204819 0.02409639Get number of unique values for numeric columns
A good way to identify categorical data within numeric columns….
mtcars %>%
dplyr::select(where(is.numeric)) %>%
purrr::map_df(~ unique(.) %>% length()) %>%
tidyr::pivot_longer(everything()) %>%
dplyr::arrange(value)
#> # A tibble: 11 x 2
#> name value
#> <chr> <int>
#> 1 vs 2
#> 2 am 2
#> 3 cyl 3
#> 4 gear 3
#> 5 carb 6
#> 6 hp 22
#> 7 drat 22
#> 8 mpg 25
#> 9 disp 27
#> 10 wt 29
#> 11 qsec 30